Mastering Michaelis-Menten Kinetics with NONMEM: A Comprehensive Guide to Parameter Estimation for Drug Development
This article provides a comprehensive guide for researchers and drug development professionals on estimating Michaelis-Menten pharmacokinetic parameters using NONMEM, the industry-standard nonlinear mixed-effects modeling software [citation:7].
Mastering Michaelis-Menten Kinetics with NONMEM: A Comprehensive Guide to Parameter Estimation for Drug Development
Abstract
This article provides a comprehensive guide for researchers and drug development professionals on estimating Michaelis-Menten pharmacokinetic parameters using NONMEM, the industry-standard nonlinear mixed-effects modeling software [citation:7]. It covers the full scope from foundational concepts of saturable elimination to advanced application, troubleshooting model instability [citation:1], and comparative analysis with emerging AI-based methodologies [citation:2]. The content details practical workflows for model implementation, addresses common convergence challenges, explores strategies for generating robust initial estimates [citation:3], and demonstrates application in therapeutic dose optimization [citation:4]. This guide synthesizes traditional best practices with the latest advancements in automated modeling pipelines to equip scientists with the knowledge to reliably characterize nonlinear pharmacokinetics.
Understanding Michaelis-Menten Kinetics and NONMEM's Role in Population PK/PD Analysis
Theoretical Foundations of Michaelis-Menten Kinetics
The Michaelis-Menten equation describes the rate of enzymatic reactions, where the velocity (v) of product formation depends on the substrate concentration ([S]) [1]. The fundamental form of the equation is: v = (Vmax × [S]) / (Km + [S]) [1] [2] [3].
The two critical parameters are:
- V_max (Maximum Reaction Velocity): The theoretical maximum rate of the reaction, achieved when all enzyme active sites are saturated with substrate [1]. In pharmacokinetics, it represents the maximum capacity of an enzyme system to metabolize a drug.
- Km (Michaelis Constant): Defined as the substrate concentration at which the reaction velocity is half of Vmax [1] [2]. It is an inverse measure of the enzyme's affinity for the substrate; a lower K_m indicates higher affinity [4].
The model is derived from the foundational enzymatic mechanism: E + S ⇌ ES → E + P where E is the free enzyme, S is the substrate, ES is the enzyme-substrate complex, and P is the product [1] [3]. The derivation assumes steady-state conditions for the ES complex or rapid equilibrium binding [1] [3].
In pharmacokinetics (PK), this model is adapted to describe nonlinear drug elimination, commonly observed with drugs like phenytoin, voriconazole, and ethanol [5]. Here, the rate of drug metabolism (or elimination) is saturable. The PK analogue of the Michaelis-Menten equation is: -dC/dt = (Vmax × C) / (Km + C) where C is the plasma drug concentration [5] [2].
Pharmacokinetic Translation and Clinical Significance
The translation of in vitro enzyme kinetics to in vivo pharmacokinetics is central to predicting drug behavior. Key derived PK equations include [5]:
- Clearance (CL): At steady state, clearance becomes concentration-dependent: CL = Vmax / (Km + C_ss).
- Dosing Rate (DR): For drugs with nonlinear kinetics, the maintenance dose rate to achieve a target steady-state concentration (Css) is: DR = (Vmax × Css) / (Km + C_ss).
When drug concentration C is much lower than Km (C << Km), the elimination approximates first-order kinetics (linear pharmacokinetics). When C is much greater than Km (C >> Km), the elimination rate approaches the constant V_max, demonstrating zero-order kinetics [1] [5]. This transition underpins the nonlinear, dose-dependent pharmacokinetics critical for drugs with a narrow therapeutic index.
The specificity constant (kcat/Km), representing the enzyme's catalytic efficiency, is crucial for understanding in vivo drug metabolism and interactions [1]. Drugs can act as enzyme inducers (increasing Vmax) or inhibitors (decreasing Vmax or increasing apparent K_m), altering their own kinetics or that of co-administered drugs [5].
Table 1: Representative Drugs Exhibiting Michaelis-Menten Pharmacokinetics
| Drug/Therapeutic Area | Primary Metabolizing Enzyme | Clinical PK Implication |
|---|---|---|
| Phenytoin (Anticonvulsant) | CYP2C9 | Dose-dependent elimination; small dose increases can lead to large rises in concentration [5]. |
| Voriconazole (Antifungal) | CYP2C19 | Nonlinear PK; therapeutic drug monitoring required [5]. |
| Ethanol | Alcohol dehydrogenase | Zero-order elimination at high concentrations [5]. |
| Theophylline (Bronchodilator) | CYP1A2 | Concentration-dependent clearance; narrow therapeutic window [5]. |
Parameter Estimation Methods & The Role of NONMEM
Accurate estimation of Vmax and Km from experimental data is paramount. Methods range from traditional linearizations to modern nonlinear mixed-effects modeling.
- Linear Transformation Methods: Historically, plots like Lineweaver-Burk (1/v vs. 1/[S]) and Eadie-Hofstee (v vs. v/[S]) were used to graphically estimate parameters [4]. These methods are simple but can distort error structures and are statistically inferior for parameter estimation [4] [6].
- Direct Nonlinear Regression: Fitting the untransformed velocity vs. concentration data directly to the Michaelis-Menten equation using nonlinear least squares regression is more robust and avoids the biases of linear transformations [4] [6].
In clinical pharmacology, population pharmacokinetic (PopPK) modeling with nonlinear mixed-effects models (NONMEM) is the gold standard for estimating Vmax and Km from sparse clinical data. It distinguishes between:
- Fixed Effects: The typical population values of Vmax and Km.
- Random Effects: Inter-individual variability (IIV) around these parameters and residual unexplained variability [7].
A major challenge in nonlinear modeling is obtaining good initial parameter estimates for the optimization algorithm. Poor initial estimates can lead to model convergence failures [7]. Strategies to generate initial estimates include [7]:
- Adaptive Single-Point Method: Using concentrations from specific time points (e.g., after first dose or at steady-state) with known dosing information to approximate clearance and volume.
- Graphical Methods: Visual inspection of concentration-time profiles.
- Naïve Pooled Data Analysis: Treating all data as if from a single individual for initial approximations.
- Parameter Sweeping: Testing a range of candidate values and selecting those with the best predictive performance [7].
Recent research has focused on developing automated pipelines that integrate these data-driven methods to generate robust initial estimates without user input, facilitating large-scale and automated PopPK analyses [7].
Table 2: Comparison of Parameter Estimation Methods for Michaelis-Menten Kinetics
| Method | Principle | Advantages | Disadvantages/Considerations |
|---|---|---|---|
| Lineweaver-Burk Plot | Linear transform: Plot 1/v vs. 1/[S]. Vmax = 1/y-intercept; Km = slope/y-intercept [4]. | Simple visualization. | Prone to error propagation; gives undue weight to low-concentration data points; statistically unreliable [4]. |
| Direct Nonlinear Regression (NONMEM) | Fits [S] vs. time data directly to the differential form of the MM equation [4]. | Most accurate and precise; handles error structure correctly; suitable for sparse, population data [4]. | Requires specialized software; computationally intensive; needs good initial estimates. |
| Eadie-Hofstee Plot | Linear transform: Plot v vs. v/[S]. Vmax = y-intercept; Km = -slope [4]. | Visualizes data spread. | Less distortion than Lineweaver-Burk but still suboptimal vs. nonlinear methods [4]. |
| Automated Pipeline (e.g., R-based) | Combines single-point, graphical, and NCA methods to generate initial estimates for PopPK [7]. | Automated, robust, applicable to rich and sparse data; reduces modeler burden [7]. | Relatively new approach; may require validation for specific model structures. |
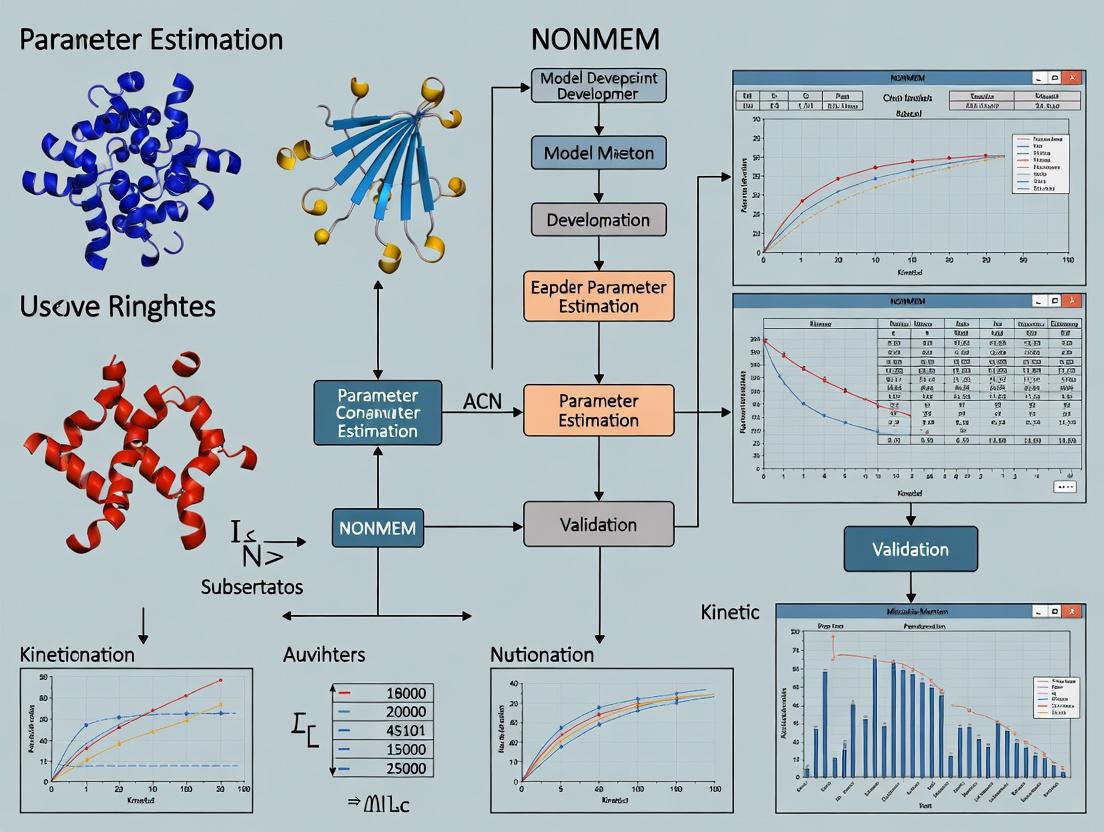
Diagram: Workflow for Michaelis-Menten Parameter Estimation in Pharmacokinetics
Advanced Protocols: NONMEM Implementation for MM Parameters
Protocol 1: Implementing a Basic Michaelis-Menten Elimination Model in NONMEM
This protocol outlines the steps to code a one-compartment model with Michaelis-Menten elimination in a NONMEM control stream ($PROBLEM, $INPUT, etc.).
- $SUBROUTINE: Select
ADVAN6orADVAN13for general differential equation solutions, orADVAN10for analytical solutions to specific nonlinear models. DefineTOLappropriately for stiff equations. - $MODEL: Define compartments (e.g.,
DEPOT,CENTRAL). $PK:
$DES (Differential Equations for
ADVAN6):$ERROR: Define an appropriate residual error model (e.g., additive, proportional, or combined).
$ESTIMATION: Use a method like
FOCE INTERACTIONfor reliable estimation of nonlinear models with random effects [8].- $COVARIANCE: Request covariance step to obtain standard errors of parameter estimates.
Protocol 2: Generating Initial Estimates for PopPK Analysis [7]
An automated R-based pipeline can generate initial estimates for THETA:
- Data Preparation: Pool data across individuals based on time after dose (TAD), calculating median concentrations in time bins.
- Estimate Half-life: Perform linear regression on the terminal phase of the pooled log-concentration vs. time data.
- Calculate Parameters:
- For intravenous data, use an early post-first-dose point (within 0.2 half-lives) to estimate volume: Vd ≈ Dose / C₁.
- Use steady-state maximum (Css,max) and minimum (Css,min) concentrations to estimate clearance: CL ≈ Dose * ln(Css,max/Css,min) / [τ * (Css,max - Css,min)], where τ is the dosing interval.
- Approximate
V_maxandK_mfrom a range of doses and steady-state concentrations using the linearized form: 1/DR = (Km/Vmax) * (1/Css) + 1/Vmax.
- Parameter Sweeping: If analytical methods are insufficient, simulate concentration-time profiles for a grid of candidate (Vmax, Km) pairs and select the pair minimizing the relative root mean squared error (rRMSE) between observed and predicted data.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Reagents & Software for MM Parameter Estimation Research
| Item/Software | Function in MM/PK Research | Example/Note |
|---|---|---|
| NONMEM | Industry-standard software for nonlinear mixed-effects modeling of PK/PD data. Used for population estimation of Vmax, Km, and their variability [4] [8]. | ICON Development Solutions. Required for implementing protocols in Sections 3 & 4. |
| R or Python | Open-source programming environments for data simulation, exploratory analysis, running automated estimation pipelines [7], and creating diagnostic plots. | Packages: nlmixr2, mrgsolve (R), PyDarwin, Pharmpy (Python). |
| ADP-Glo Kinase Assay | A luminescent method to measure kinase activity by quantifying ADP production. Used to generate initial velocity (v) vs. substrate concentration ([S]) data for in vitro enzyme kinetic studies [6]. | Example from c-Src kinase inhibition studies [6]. |
| High-Throughput Microplate Reader | Instrument to measure absorbance, fluorescence, or luminescence in 96- or 384-well plates. Essential for rapidly collecting the dense [S]-v data required for robust in vitro MM parameter fitting [6]. | GloMax Discover (Promega) [6]. |
| GraphPad Prism | Commercial software for scientific graphing and basic nonlinear regression. Useful for initial exploratory fitting of in vitro MM kinetics and inhibitor IC₅₀/Kᵢ calculations [6]. | Commonly used before advanced PopPK analysis in NONMEM. |
| SigmaPlot/Solver (Excel) | Software for nonlinear curve fitting. Can be used to fit data directly to the MM equation to obtain preliminary Vmax and Km estimates [5] [6]. | An accessible tool for direct nonlinear regression [6]. |
Saturable elimination, also known as zero-order or capacity-limited elimination, is a critical pharmacokinetic (PK) phenomenon where drug clearance pathways become saturated at higher concentrations, leading to non-proportional increases in systemic exposure with dose escalation [9]. This occurs when the enzymes responsible for metabolism (e.g., cytochrome P450 isoforms) or active transport processes reach their maximum catalytic capacity (Vmax) [10]. In contrast, first-order elimination, which governs most drugs at therapeutic doses, describes a process where a constant fraction of drug is eliminated per unit time [9].
The transition from first-order to zero-order kinetics has profound implications for drug safety and efficacy. When elimination is saturated, small increases in dose can lead to disproportionately large increases in drug exposure (area under the curve, AUC), dramatically elevating the risk of toxicity [9]. This is a cornerstone of nonlinear pharmacokinetics and is classically described by the Michaelis-Menten (M-M) equation [10].
Understanding and characterizing saturable elimination is therefore essential across all drug modalities. For small molecules, saturation often involves metabolic enzymes like CYP2C9, CYP2D6, or CYP3A4. For biologics such as monoclonal antibodies, saturation can occur via target-mediated drug disposition (TMDD), where the high-affinity binding to a pharmacological target acts as a saturable elimination pathway [11]. Accurately estimating the parameters of the M-M equation (Vmax and Km) is thus a fundamental task in model-informed drug development (MIDD), enabling the prediction of safe and effective dosing regimens across populations [11].
This application note details the principles, experimental strategies, and advanced analytical methodologies—particularly using NONMEM for parameter estimation—to study saturable elimination throughout the drug development lifecycle.
Quantitative Characterization of Elimination Kinetics
The fundamental difference between linear and saturable elimination is quantitatively distinct, impacting all key pharmacokinetic parameters. The following table contrasts the characteristics of both kinetic processes [12] [9].
Table 1: Characteristics of First-Order vs. Zero-Order (Saturable) Elimination Kinetics
| Pharmacokinetic Parameter | First-Order (Linear) Elimination | Zero-Order (Saturable) Elimination |
|---|---|---|
| Governing Principle | A constant fraction of drug is eliminated per unit time. | A constant amount of drug is eliminated per unit time. |
| Rate of Elimination | Proportional to drug concentration (Rate = ke × [C]). | Constant, independent of drug concentration (Rate = Vmax). |
| Mathematical Description | dC/dt = -ke × C | dC/dt = -Vmax |
| Plasma Concentration-Time Profile | Exponential decay. Linear on a semi-log plot. | Linear decay. |
| Half-life (t½) | Constant, independent of dose. | Increases with increasing dose/concentration. |
| Area Under the Curve (AUC) | Increases proportionally with dose. | Increases more than proportionally with dose. |
| Time to Eliminate Drug | Independent of dose (~4-5 half-lives). | Dependent on dose; longer with higher doses. |
| Clinical Dose Predictability | Highly predictable; doubling dose doubles exposure. | Unpredictable; small dose increases can cause large exposure spikes. |
| Common Examples | Most drugs at therapeutic doses (e.g., amoxicillin, lorazepam). | Phenytoin, salicylates (high dose), ethanol, paclitaxel [9]. |
The Michaelis-Menten equation bridges these concepts, describing the velocity (V) of an enzymatic reaction or saturable process as a function of substrate concentration ([S]) [10]:
V = (Vmax × [S]) / (Km + [S])
Where:
- Vmax is the maximum elimination rate (capacity).
- Km (Michaelis constant) is the substrate concentration at half of Vmax, representing the affinity of the enzyme/transporter for the substrate.
At very low concentrations ([S] << Km), the equation simplifies to V ≈ (Vmax/Km) × [S], exhibiting first-order kinetics. At high concentrations ([S] >> Km), V ≈ Vmax, and elimination becomes zero-order and saturable [10] [9].
Protocols for Characterizing Saturable Elimination
Protocol for In Vitro Michaelis-Menten Parameter Estimation Using NONMEM
Adapted from the simulation study by Cho & Lim (2018) [10] [13].
Objective: To determine the most accurate and precise method for estimating Vmax and Km from in vitro enzyme kinetic data, comparing traditional linearization methods with nonlinear estimation in NONMEM.
Materials & Software:
- R Statistical Software (with
deSolvepackage) - NONMEM (Version 7.3 or higher)
- Simulated or experimental time-course substrate concentration data.
Procedure:
Data Generation/Collection:
- For simulation studies: Generate substrate depletion time-course data for multiple initial substrate concentrations (e.g., 20.8, 41.6, 83, 166.7, 333 mM) using the M-M differential equation:
d[S]/dt = - (Vmax × [S]) / (Km + [S]). Incorporate realistic residual error models (additive, proportional, or combined) [10]. - For experimental data: Conduct in vitro metabolic stability assays with human liver microsomes or recombinant enzymes, measuring substrate loss over time at 5-6 different starting concentrations.
- For simulation studies: Generate substrate depletion time-course data for multiple initial substrate concentrations (e.g., 20.8, 41.6, 83, 166.7, 333 mM) using the M-M differential equation:
Data Preparation for Different Estimation Methods:
- NM (NONMEM Nonlinear): Use the raw [S]-time data directly.
- LB (Lineweaver-Burk) & EH (Eadie-Hofstee): Calculate initial velocity (Vi) for each concentration from the linear slope of the early time points. Transform data: LB uses
1/Vi vs. 1/[S]; EH usesVi vs. Vi/[S][13]. - NL (Nonlinear Vi-[S]): Use the calculated Vi and corresponding [S] pair.
- ND (Nonlinear Delta): Calculate the average rate between adjacent time points (VND) and the midpoint concentration ([S]ND) [13].
Parameter Estimation in NONMEM:
- Code the appropriate structural model for each method in NONMEM control files.
- For method NM, implement the differential equation directly.
- For methods LB and EH, use linear models on the transformed data. For NL and ND, use the M-M algebraic equation.
- Use the First-Order Conditional Estimation (FOCE) method with interaction.
- Perform estimation for all datasets (simulated replicates or experimental runs).
Analysis & Comparison:
- For each method, compile the estimated Vmax and Km values.
- Calculate measures of accuracy (e.g., median relative error) and precision (e.g., 90% confidence interval width) relative to the known true values (simulation) or the most robust estimate.
- Expected Outcome: As demonstrated by Cho & Lim, nonlinear regression fitting the full [S]-time data (NM method) typically provides the most accurate and precise parameter estimates, especially with complex error structures, outperforming traditional linearization methods (LB, EH) [10].
Protocol for Identifying Saturable Elimination in Phase I Clinical Data Using Machine Learning
Informed by Certara's application of machine learning for PK/PD modeling [14].
Objective: To efficiently identify the structural PK model—including the potential for saturable (Michaelis-Menten) elimination—that best describes complex Phase I clinical trial data.
Materials & Software:
- Phase I rich concentration-time data.
- Machine learning-enabled pharmacometric software (e.g., containing genetic algorithm capabilities).
- Standard NONMEM/Pirana for final model refinement.
Procedure:
Exploratory Data Analysis (EDA):
- Plot individual patient concentration-time profiles.
- Plot AUC vs. Dose. A greater-than-proportional increase suggests saturable elimination or absorption [15].
- Plot clearance (CL) vs. concentration. A decreasing trend with higher concentrations suggests saturable elimination.
Define the Model Space:
- Define a set of plausible structural models to be tested. This is a critical step driven by the scientist.
- Absorption: First-order with/without lag time, sequential zero-first order, distributed delay function.
- Distribution: 1-, 2-, or 3-compartment models.
- Elimination: Linear (first-order) clearance vs. Michaelis-Menten (saturable) clearance.
- Covariates: Include potential relationships (e.g., weight on volume, age on clearance).
Machine Learning Model Search:
- Configure a genetic algorithm or other global search algorithm to explore the defined model space.
- The algorithm will automatically run hundreds to thousands of model configurations, evaluating each based on a fitness score (e.g., objective function value penalized for number of parameters) [14].
- The search is non-sequential, evaluating combinations of features (e.g., dual-compartment with saturable elimination) that might be missed in manual stepwise building.
Model Evaluation and Selection:
- Review the top-performing models identified by the ML algorithm.
- Perform standard pharmacometric diagnostics on the leading candidates: goodness-of-fit plots, visual predictive checks, precision of parameter estimates.
- Final Selection: The scientist makes the final decision based on statistical fit, parsimony, and biological plausibility. The ML tool provides robust, data-driven candidates [14].
Diagram 1: Workflow for Identifying Saturable Kinetics in Clinical Data
The Scientist's Toolkit: Essential Reagents & Software
Table 2: Key Research Reagent Solutions for Studying Saturable Elimination
| Tool/Reagent Category | Specific Examples | Function in Saturable Elimination Research |
|---|---|---|
| In Vitro Metabolism Systems | Human liver microsomes (HLM), recombinant CYP enzymes, hepatocytes. | Provide the biological enzymes to conduct in vitro kinetic studies for estimating initial Vmax and Km values for small molecules. |
| Transport Assay Systems | Transfected cell lines (e.g., MDCK, HEK293) overexpressing specific transporters (OATP1B1, P-gp). | Characterize saturable active transport processes that may contribute to uptake or elimination. |
| Target Proteins (for Biologics) | Soluble recombinant target antigens, cell lines expressing membrane-bound targets. | Essential for characterizing the target-binding affinity (KD) and capacity (Rmax) in TMDD assays, a key saturable pathway for biologics. |
| Simulation & Modeling Software | R (with deSolve, nlmixr), NONMEM, Monolix, Phoenix NLME. |
The primary platform for nonlinear mixed-effects modeling, enabling population estimation of M-M parameters from clinical data. |
| Global Optimization Algorithms | Particle Swarm Optimization (PSO), Genetic Algorithms (GA). | Address challenges in nonlinear model fitting, such as parameter identifiability and convergence, by searching the global parameter space [16]. |
| Specialized NONMEM Extensions | P-NONMEM [16], Fractional Differential Equation (FDE) subroutines [17]. | Extend NONMEM's capability for complex models: PSO integration for robust estimation [16]; FDEs for modeling anomalous "power-law" kinetics sometimes linked to nonlinear elimination [17]. |
| Machine Learning Platforms for MIDD | Certara's ML tools, custom Python/R scripts using TensorFlow/Scikit-learn. | Automate the exploration of complex PK model spaces to efficiently identify the inclusion of saturable elimination mechanisms [14]. |
Advanced Analytical Considerations & NONMEM Workflow
Addressing Parameter Identifiability in Nonlinear Models
A major challenge in estimating M-M parameters (Vmax, Km) from in vivo data is statistical non-identifiability, where multiple parameter combinations yield an equally good fit to the data [16]. This often leads to estimation failures or unstable results in NONMEM.
Protocol Mitigation Strategies:
- Informative Prior Design: Collect rich data across a wide dose range, especially with doses designed to produce concentrations near and above the estimated Km.
- Use of Global Optimizers: Implement hybrid algorithms like LPSO (Particle Swarm Optimization coupled with local search) within or alongside NONMEM. As derivative-free methods, they are less prone to failure from singularity issues and can better navigate complex objective function landscapes to find global solutions [16].
- Model Simplification: If Vmax and Km are not jointly identifiable, consider fixing one parameter to a literature value from in vitro studies (e.g., Km) and estimating the other (Vmax in vivo).
NONMEM Workflow for Population M-M Parameter Estimation
Diagram 2: NONMEM Workflow for Population Michaelis-Menten Analysis
Application Across Modalities: Small Molecules vs. Biologics
The manifestation and analysis of saturable elimination differ between drug classes:
Small Molecules: Saturable elimination is primarily metabolism-driven. The focus is on characterizing the specific enzymes involved (e.g., via chemical inhibition assays in vitro) and applying the standard M-M framework. Phenytoin and high-dose aspirin are classic examples [9]. The NONMEM model typically adds a Michaelis-Menten clearance term:
CL = (Vmax / (Km + C)).Biologics (e.g., mAbs): Saturable elimination is often target-mediated (TMDD). At low doses, the drug binds with high affinity to its target, and the drug-target complex is internalized and degraded, creating a high-capacity, saturable elimination pathway. At higher doses, the target is saturated, and linear, non-specific clearance pathways dominate [11]. Modeling is more complex, often requiring a quasi-equilibrium or quasi-steady-state approximation of the full TMDD model within NONMEM, which still relies on estimating a saturation constant (Km) related to target affinity and turnover.
In both cases, the accurate estimation of saturation parameters is critical for first-in-human dose prediction, optimizing dosing regimens for Phase II/III, and predicting drug-drug interaction potential (for small molecules) [11].
NONMEM (Nonlinear Mixed Effects Model) is a computer program written in FORTRAN 90/95, designed for fitting general nonlinear regression models to data, with specialized application in population pharmacokinetics (PK) and pharmacodynamics (PD) [18] [19]. Developed by Lewis Sheiner and Stuart Beal at the University of California, San Francisco, it has become the industry-standard software for population PK/PD analysis for over 30 years [20] [21]. The software is particularly powerful for analyzing data with sparse sampling or variable designs across individuals, as it accounts for both inter-individual (random effects) and intra-individual variability, as well as measured covariates (fixed effects) [18] [19].
The NONMEM system comprises three core components [20] [18]:
- NONMEM: The foundational, general nonlinear regression program.
- PREDPP (PRED for Population Pharmacokinetics): A specialized subroutine library that handles standard and complex kinetic models, freeing users from coding common differential equations. It includes a suite of
ADVANmodel subroutines [19]. - NM-TRAN (NONMEM Translator): A preprocessor that translates user-friendly control streams and data files into the formats required by NONMEM [20] [18].
The current version, NONMEM 7.6, includes advanced estimation methods such as Stochastic Approximation Expectation-Maximization (SAEM) and Markov-Chain Monte Carlo Bayesian Analysis (BAYES, NUTS), along with support for complex mathematical constructs like delay differential equations (ADVAN16, ADVAN17) and distributed delay convolutions [20]. It also supports parallel computing to reduce run times for large problems [20].
The following diagram illustrates the workflow and relationship between these core components and the user's inputs.
NONMEM System Architecture and Workflow
Core Components and Model Library
NONMEM's power for pharmacometric analysis is largely enabled by the PREDPP library. Users select a pre-defined model type via ADVAN subroutines in their control stream. For research focused on Michaelis-Menten kinetics, ADVAN10 is directly applicable as it implements a one-compartment model with Michaelis-Menten elimination [19]. Other ADVAN subroutines support a wide range of linear and nonlinear PK/PD models.
Table: Selected PREDPP (ADVAN) Model Subroutines [19]
| Model Subroutine | Compartments | Basic Parameters | Model Description |
|---|---|---|---|
| ADVAN1 | 1 Central, 1 Output | K, V | One-compartment linear model. |
| ADVAN2 | 1 Depot, 1 Central, 1 Output | KA, K, V | One-compartment model with first-order absorption. |
| ADVAN3 | 1 Central, 1 Peripheral, 1 Output | K, K12, K21 | Two-compartment linear mammillary model. |
| ADVAN4 | 1 Depot, 1 Central, 1 Peripheral, 1 Output | KA, K, K23, K32 | Two-compartment model with first-order absorption. |
| ADVAN10 | 1 Central, 1 Output | VM, KM | One-compartment model with Michaelis-Menten elimination. |
| ADVAN13 | General Nonlinear | User-defined | General nonlinear model with stiff/non-stiff ODE solver (LSODA). |
The Scientist's Toolkit: Essential Research Reagents and Materials
For conducting NONMEM-based Michaelis-Menten research, the following components are essential.
Table: Essential Toolkit for NONMEM-based Research
| Item | Function / Description | Key Considerations |
|---|---|---|
| NONMEM Software | The core estimation engine for performing nonlinear mixed-effects modeling [20]. | Licensed annually from ICON plc. Version 7.6 is the latest [20]. |
| Fortran Compiler | Required to compile NONMEM's source code into an executable program [20] [22]. | Intel Fortran Compiler 9.0+ or gFortran for Windows/Linux are common [20]. |
| Control Stream File (.ctl) | A user-written text file containing all modeling instructions, commands, and model code for NM-TRAN [19] [22]. | Defines the structural model ($SUB, $PK), error model ($ERROR), estimation method ($EST), and requested output ($TABLE). |
| Data File (.csv or .prn) | A text file containing the experimental or clinical data for analysis [19] [22]. | Must include required data items (e.g., ID, TIME, DV, AMT) in a space- or comma-delimited format. |
| Interface/Manager | Third-party tools that facilitate project management, run execution, and graphical diagnostics [19]. | Examples include Pirana, Wings for NONMEM (WFN), and Perl Speaks NONMEM (PsN). |
| Graphical Diagnostic Tool | Software for generating diagnostic plots (e.g., goodness-of-fit, VPC) from NONMEM output tables [23]. | R (with packages like xpose), Xpose, or custom scripts in Python/MATLAB. |
Application to Michaelis-Menten Parameter Estimation
The Michaelis-Menten equation (V = (Vmax * [S]) / (Km + [S])) is fundamental for characterizing enzyme kinetics and saturable elimination processes in drug metabolism [24]. Accurate estimation of its parameters, the maximum reaction rate (Vmax) and the Michaelis constant (Km), is critical for in vitro to in vivo extrapolation and predicting nonlinear PK.
Traditional linearization methods (e.g., Lineweaver-Burk, Eadie-Hofstee) are prone to statistical bias because they distort the error structure of the data. Nonlinear mixed-effects modeling with NONMEM provides a more robust framework by directly fitting the nonlinear model to the data, properly handling both fixed effects (typical parameter values) and random effects (inter-experiment/inter-individual variability) [24].
Comparative Performance of Estimation Methods
A key simulation study directly compared the performance of various estimation methods for Michaelis-Menten parameters [24]. The results unequivocally support the use of nonlinear methods in NONMEM.
Table: Performance Comparison of Michaelis-Menten Parameter Estimation Methods [24]
| Estimation Method | Category | Relative Accuracy & Precision for Vmax and Km | Key Finding |
|---|---|---|---|
| Nonlinear Method (NM) in NONMEM | Nonlinear, Mixed-Effects | Most accurate and precise | Provides the most reliable parameter estimates by correctly specifying the error model. |
| Traditional Linearization Methods (e.g., Lineweaver-Burk) | Linear, Transform-based | Less accurate and precise | Amplifies error and distorts variance, leading to biased parameter estimates. |
| Iterative Two-Stage (ITS) | Hybrid, Nonlinear | Intermediate performance | More robust than linearization but generally less efficient than full population methods like FOCE or SAEM. |
The study found that the superiority of NONMEM's nonlinear estimation was particularly evident when data followed a combined (proportional plus additive) error model, which is common in real-world biological data [24]. This makes NONMEM the preferred tool for reliable Michaelis-Menten parameter estimation in pharmacological research.
Experimental Protocols for Michaelis-Menten Analysis
This section provides detailed, actionable protocols for setting up and executing a NONMEM analysis aimed at estimating Michaelis-Menten parameters, as informed by simulation studies and tutorial guides [24] [23].
Protocol 1: Simulation-Based Validation of Method Performance
Objective: To validate and compare the accuracy of different estimation methods for Vmax and Km using simulated in vitro elimination kinetic data [24].
Define True Parameters & Model:
- Set population values for
Vmax(e.g., 100 nmol/min/mg) andKm(e.g., 10 µM). - Define an error model: combined additive (
σ_add) and proportional (σ_prop) error is recommended (e.g.,DV = IPRED * (1 + ε₁) + ε₂, whereε₁andε₂are normally distributed with mean 0 and variancesσ_prop²andσ_add²).
- Set population values for
Generate Simulation Dataset:
- Using software like R, generate substrate concentrations (
[S]) over a range (e.g., 0.2Km to 5Km). - For each
[S], calculate the true reaction velocity (V) using the Michaelis-Menten equation. - Add random noise to
Vbased on the defined error model to create observed values (DV). - Replicate this process to create
N(e.g., 1000) simulated datasets [24].
- Using software like R, generate substrate concentrations (
Prepare NONMEM Data File:
- Structure a .csv file with columns:
SIM,ID,TIME,DV,CONC. UseTIMEas a placeholder if not time-course data;CONCholds the substrate concentration ([S]).
- Structure a .csv file with columns:
Develop NONMEM Control Stream:
- Use
$SUBROUTINE ADVAN10 TOL=6for Michaelis-Menten kinetics. - In
$PK: DefineVM = THETA(1) * EXP(ETA(1))andKM = THETA(2) * EXP(ETA(2)). Set initialTHETAvalues close to the true simulated values. - In
$ERROR: DefineIPREDusing the Michaelis-Menten equation with model parameters, and defineY = IPRED + IPRED*EPS(1) + EPS(2)for a combined error model. - Use an estimation method like
FOCEwithINTERACTIONin$ESTIMATION.
- Use
Execute and Analyze:
- Run NONMEM for each simulated dataset or use the
$SIMULATIONfunctionality. - For each run, record the estimated
VmaxandKm. - Calculate performance metrics (e.g., relative bias, precision) across all simulations to compare with other methods [24].
- Run NONMEM for each simulated dataset or use the
Protocol 2: Population Analysis of Enzyme Kinetic Data
Objective: To estimate typical population values and inter-assay variability of Vmax and Km from a real in vitro experiment conducted with multiple enzyme preparations (e.g., different hepatocyte lots) [23].
Data Assembly and Exploration:
- Compile data from all experiments. Essential data items:
ID(experiment/lot number), substrate concentration (CONCorSSas a steady-state indicator), and observed reaction velocity (DV). - Perform exploratory graphical analysis (velocity vs. concentration) to check for saturable kinetics and outliers.
- Compile data from all experiments. Essential data items:
Create NONMEM Data File:
- Format data as a space- or comma-delimited text file. Map columns using
$INPUTin the control stream (e.g.,$INPUT ID TIME DV CONC AMT). UseAMT=0for observation records.
- Format data as a space- or comma-delimited text file. Map columns using
Base Model Development:
- Control Stream Setup:
$PROBLEMPopulation Michaelis-Menten Kinetics$DATAyour_data.csvIGNORE=@$SUBROUTINE ADVAN10 TOL=6$ABBR(Optional, for advanced reparameterization)
- Parameter Definition (
$PK):- Define typical population parameters:
TVVM = THETA(1)andTVKM = THETA(2). - Define inter-individual variability (IIV) using exponential error models:
VM = TVVM * EXP(ETA(1))andKM = TVKM * EXP(ETA(2)). - Initial
THETAestimates can come from prior literature or graphical estimates.
- Define typical population parameters:
- Error Model (
$ERROR):- Define the individual prediction:
IPRED = (VM * CONC) / (KM + CONC). - Choose a residual error model. A proportional error is often suitable:
Y = IPRED + IPRED*EPS(1).
- Define the individual prediction:
- Estimation and Output:
$ESTIMATION METHOD=FOCE INTERACTION MAXEVAL=9999 SIGDIGITS=3$TABLE ID TIME IPRED PRED RES WRES CWRES NOPRINT ONEHEADER FILE=sdtab1$TABLE ID ETA1 ETA2 NOPRINT ONEHEADER FILE=patab1
- Control Stream Setup:
Model Evaluation:
- Run the model and assess convergence (successful covariance step).
- Generate diagnostic plots (e.g., PRED vs. DV, CWRES vs. PRED) using the output tables to evaluate goodness-of-fit [23].
Covariate Model Building (Optional):
- If experiment metadata is available (e.g., protein content, donor age), test for covariate relationships on
VmaxorKmin the$PKblock using linear or power functions (e.g.,TVVM = THETA(1) * (PROT/MEANPROT)THETA(3)). - Use objective function value (OFV) changes and diagnostic plots to guide model selection.
- If experiment metadata is available (e.g., protein content, donor age), test for covariate relationships on
The logical process for developing and refining a population Michaelis-Menten model is summarized in the diagram below.
Population Michaelis-Menten Model Development Workflow
Advanced Features and Future Directions
NONMEM 7.6 includes sophisticated features that extend its utility beyond standard PK/PD modeling, several of which are relevant for complex kinetic analyses [20] [25].
- Delay Differential Equations (DDEs): Modeled via
ADVAN16andADVAN17, these are essential for capturing physiological delays (e.g., in cell proliferation or indirect response models) [20] [25]. - Nonparametric and Bayesian Methods: The software supports nonparametric estimation of parameter distributions, which is valuable when random effects are non-normal [26]. Bayesian analysis (
BAYESandNUTSsampling) allows for formal incorporation of prior knowledge [20]. - Cutting-Edge Research: Recent extensions demonstrate NONMEM's adaptability, such as the implementation of fractional differential equation (FDE) models to describe anomalous kinetics or power-law behavior, which can offer more parsimonious models for complex data [17].
NONMEM remains the definitive software for nonlinear mixed-effects modeling in drug development and basic pharmacological research. Its rigorous framework for population parameter estimation, exemplified by its superior performance in estimating Michaelis-Menten constants compared to linearization methods, provides scientists with reliable and interpretable results [24]. The structured protocols for simulation and analysis, combined with its advanced capabilities for handling complex biological processes, ensure that NONMEM will continue to be an indispensable tool for quantifying and predicting nonlinear kinetics in therapeutic research.
Abstract Integrating Michaelis-Menten (MM) kinetics into a Nonlinear Mixed Effects (NLME) modeling framework is essential for accurately characterizing the saturable, non-linear elimination of numerous drugs. This integration involves defining a structural pharmacokinetic (PK) model with MM parameters (Vmax, Km), distinguishing population-level fixed effects from inter-individual random effects, and quantifying residual unexplained variability. Within software like NONMEM, this allows for the analysis of sparse, real-world clinical data to estimate typical population parameters, identify influential covariates, and quantify variability, thereby informing optimized dosing strategies. This application note provides detailed protocols and methodologies for the successful implementation and evaluation of population PK models incorporating MM elimination, framed within a broader thesis on advanced parameter estimation using NONMEM.
1. Introduction: Thesis Context and Rationale This work is situated within a broader thesis investigating robust methodologies for parameter estimation in NONMEM, with a focus on complex, non-linear kinetics. Michaelis-Menten elimination is a fundamental non-linear process where the elimination rate approaches a maximum velocity (Vmax) as concentration increases, with Km representing the concentration at half Vmax. In population modeling, the goal is to estimate the typical values (fixed effects) of Vmax and Km for a population, understand how they vary between individuals (random effects), and account for residual error [27]. This is formally executed within an NLME framework, which simultaneously analyzes data from all individuals, making it uniquely powerful for sparse clinical data [28]. Accurately defining and diagnosing these components—fixed effects, random effects (hierarchically modeled as inter-individual, inter-occasion), and residual error—is critical for developing a model that is both biologically plausible and statistically sound, ultimately supporting drug development decisions from first-in-human studies through to personalized dosing [28] [29].
2. Foundational Components of the NLME-MM Framework A population PK model integrating MM kinetics is composed of interconnected structural, statistical, and covariate sub-models.
- 2.1. Structural (PK) Model: The core kinetic model describes the typical concentration-time profile. For a one-compartment model with intravenous bolus administration and MM elimination, the differential equation is: dA/dt = - (Vmax * C) / (Km + C), where A is amount and C is concentration (C = A/V). Vmax and Km are the key structural parameters to be estimated [30] [31].
- 2.2. Statistical Model: This defines the variability components.
- Fixed Effects (θ): The typical population values for parameters (e.g., TVVmax, TVKm).
- Random Effects:
- Inter-individual Variability (IIV): Modeled using a log-normal distribution to ensure positivity. For example,
Vmaxᵢ = TVVmax * exp(ηᵢ_Vmax), where ηᵢVmax is a random variable from a normal distribution with mean 0 and variance ω²Vmax [27] [30]. - Inter-occasion Variability (IOV): Accounts for variability within an individual across different dosing occasions [32].
- Residual Unexplained Variability (RUV): Discrepancy between individual model predictions and observed concentrations. Common models include additive (
Cobs = Cpred + ε), proportional (Cobs = Cpred * (1 + ε)), or combined error structures [27].
- Inter-individual Variability (IIV): Modeled using a log-normal distribution to ensure positivity. For example,
- 2.3. Covariate Model: Explains IIV by relating parameters to patient characteristics (e.g., weight, genotype, renal function). A covariate relationship on Vmax, for instance, can be linear (
TVVmax = θ₁ + θ₂*(WT/70)) or allometric (TVVmax = θ₁ * (WT/70)^θ₂) [30]. The inclusion of the ALDH2 genotype on volume of distribution in an alcohol PK model is a specific example of a genetic covariate [31].
3. Protocol: Implementing a Population MM Model in NONMEM
- 3.1. Data Assembly and Preparation
- Dataset Structure: Prepare a NONMEM-compliant dataset with required columns:
ID,TIME,AMT,DV(dependent variable, e.g., concentration),CMT(compartment number),EVID(event identifier), and covariates (e.g.,WT,AGE,GENO). Ensure accurate handling of dosing records and observations [33]. - Data Quality: Perform graphical exploratory data analysis. Identify and document records below the limit of quantification (BLQ). Modern methods (e.g., M3 method) that incorporate the likelihood of BLQ observations are preferred over simple imputation (e.g., LLOQ/2) [27] [32].
- Dataset Structure: Prepare a NONMEM-compliant dataset with required columns:
- 3.2. Model Development and Estimation Strategy
- Base Model Building:
- Start with a structural PK model (e.g., 1- or 2-compartment) with linear elimination to establish a baseline.
- Introduce MM elimination in place of linear clearance. This often requires switching to an ADVAN13 (general differential equation) subroutine in NONMEM unless a built-in MM option is available.
- Add IIV to key parameters (typically on Vmax and Km). Use an exponential error model. Estimate the OMEGA variance-covariance matrix.
- Test different residual error models (additive, proportional, combined).
- Parameter Estimation: Choose an appropriate estimation method. The First Order Conditional Estimation with interaction (FOCEI) is often a robust starting point. For complex MM models or datasets with high IIV/RUV, more advanced methods like Stochastic Approximation Expectation Maximization (SAEM) or Importance Sampling (IMP) may be required for stability and accuracy [32]. Bayesian Markov Chain Monte Carlo (MCMC) methods are also applicable, especially with informative priors [31].
- Covariate Analysis:
- Stepwise Forward Inclusion: Test plausible covariate-parameter relationships (e.g., weight on Vmax, creatinine clearance on Km) using likelihood ratio tests (LRT). A drop in objective function value (OFV) > 3.84 (χ², p<0.05, 1 d.f.) suggests significance.
- Backward Elimination: Refine the full model by removing non-significant covariates (increase in OFV < 6.63, p<0.01, 1 d.f.) to develop a final parsimonious model [27].
- Base Model Building:
- 3.3. Model Evaluation and Validation
- Goodness-of-Fit (GOF): Assess basic GOF plots: Observations (DV) vs. Population Predictions (PRED), DV vs. Individual Predictions (IPRED), and Conditional Weighted Residuals (CWRES) vs. TIME or PRED [32].
- Visual Predictive Check (VPC): The gold standard for model evaluation. Simulate 1000 datasets using the final model parameter estimates and overlay the observed data percentiles with the simulated prediction intervals. This assesses the model's predictive performance across the entire concentration-time profile [17].
- Numerical Predictive Check (NPC): Provides a quantitative summary of the VPC.
- Bootstrap: Perform a non-parametric bootstrap (e.g., 1000 samples) to assess the robustness and precision of the final parameter estimates and generate confidence intervals.
4. Data Presentation: Key Parameter Estimates and Model Diagnostics Table 1: Comparison of NONMEM Estimation Methods for NLME-MM Models
| Estimation Method | Key Principle | Advantages | Disadvantages | Suitability for MM Models |
|---|---|---|---|---|
| FO (First Order) | Linearizes the random effects model. | Fast, stable. | Can produce biased estimates with high IIV or RUV. | Poor; not recommended for final MM models. |
| FOCE/I | Conditional estimation; linearizes residual error. | Generally robust, accurate for many problems. | May struggle with highly non-linear models or sparse data. | Good standard choice. |
| SAEM | Stochastic sampling of the random effects space. | Accurate for complex, highly non-linear models. | Computationally intensive, results have stochastic variability. | Very good for complex MM models. |
| Bayesian (MCMC) | Uses prior distributions combined with data likelihood. | Incorporates prior knowledge; useful for sparse data. | Choice of priors influences results; computationally intensive. | Excellent when informative priors exist (e.g., from in vitro Vmax/Km) [32] [31]. |
Table 2: Example Covariate Relationships in a Structural Parameter Model
| Parameter (P) | Covariate Model Form | NONMEM Code Snippet (Example) | Biological Interpretation |
|---|---|---|---|
| Vmax | Allometric scaling by weight | TVVMAX = THETA(1) * (WT/70)THETA(2) |
Metabolic capacity scales with body size. |
| Km | Linear influence of age | TVKM = THETA(3) + THETA(4)*(AGE-40) |
Affinity of the enzyme may change with age. |
| Vd | Influence of genotype (indicator variable) | TVV = THETA(5) * (1 + THETA(6)*GENO) |
GENO=1 for variant allele, affecting distribution [31]. |
Table 3: Example Final Parameter Estimates from a Population MM Analysis (Hypothetical Data)
| Parameter | Description | Population Estimate (RSE%) | Inter-Individual Variability (%CV) |
|---|---|---|---|
| TVVmax (mg/h) | Typical max elimination rate | 10.5 (5) | 30% |
| TVKm (mg/L) | Typical Michaelis constant | 2.1 (12) | 45% |
| TVV (L) | Typical volume of distribution | 35.0 (4) | 25% |
| θₐₗₗₒₘ | Allometric exponent on Vmax | 0.75 (Fixed) | - |
| Prop. Err (%) | Proportional residual error | 15 (10) | - |
5. Experimental Protocol: Case Study on Alcohol PK with MM Elimination
- 5.1. Aim: To develop a population PK model for alcohol incorporating Michaelis-Menten elimination and to identify significant covariates (e.g., body weight, ALDH2 genotype) [31].
- 5.2. Background: Alcohol exhibits capacity-limited metabolism via alcohol dehydrogenase. Data from 34 healthy Japanese subjects with sparse sampling times were available.
- 5.3. Experimental Methodology:
- Data: Collected blood alcohol concentration-time data after controlled administration. Covariates included demographic data and ALDH2 genotype.
- Software: Analysis performed using NONMEM 7.3 with MCMC Bayesian estimation.
- Model Building: a. Structural Model: A one-compartment model with zero-order input (absorption) and Michaelis-Menten elimination was selected. b. Statistical Model: IIV was added to Vmax, Km, and Vd using exponential models. An informative prior distribution for Km was incorporated from an external study to stabilize estimation due to limited data in the later elimination phase. c. Covariate Model: Covariates (weight, age, genotype) were tested on all parameters using a Bayesian framework, evaluating changes in posterior distributions.
- Model Evaluation: Conducted via posterior predictive checks and examination of Bayesian diagnostics (e.g., trace plots for convergence).
- 5.4. Key Findings: The final model estimated a typical Vd of 49.3 L. A significant covariate effect was found, with ALDH21/2 genotype associated with a 20.4 L lower Vd compared to the ALDH21/1 genotype. The use of Bayesian priors allowed for stable parameter estimation despite data limitations [31].
6. Visualizing the Framework and Workflow
Diagram 1: Workflow for Developing a Population PK Model with MM Kinetics.
Diagram 2: Hierarchical Relationship in an NLME-MM Model.
7. The Scientist's Toolkit: Essential Resources for NLME-MM Analysis Table 4: Key Research Reagent Solutions for Population MM Modeling
| Tool/Resource | Function/Purpose | Example/Notes |
|---|---|---|
| NONMEM Software | Industry-standard software for NLME modeling. Implements estimation algorithms (FO, FOCE, SAEM, MCMC). | Required for executing the modeling protocols described herein [32] [29]. |
| PDx-Pop / Pirana | Interface and workflow manager for NONMEM. Facilitates model run management, covariate screening, and basic graphics. | Greatly improves efficiency and reproducibility of modeling projects. |
| R / RStudio with Packages | Statistical computing environment for data preparation, advanced graphics (ggplot2), model diagnostics (xpose), and simulation. | Essential for creating VPCs, custom GOF plots, and conducting bootstraps [17]. |
| Perl Speaks NONMEM (PsN) | Perl-based toolkit for NONMEM. Automates key tasks like VPC, bootstrap, and stepwise covariate modeling (SCM). | Critical for robust model evaluation and automated workflows. |
| High-Performance Computing (HPC) Cluster | Parallel processing resource. | Significantly reduces runtime for intensive methods like SAEM, IMP, or large bootstraps. |
| Curated PK/PD Dataset | Clean, well-structured dataset compliant with NONMEM format. | Must include accurate dosing records, concentration data, and covariates. The foundation of any analysis [27] [33]. |
| In Vitro Enzyme Kinetic Data | Prior estimates of Vmax and Km from preclinical studies. | Can be used to inform Bayesian prior distributions in clinical model development, stabilizing estimation [31]. |
This document details the essential prerequisites and methodologies for conducting robust Michaelis-Menten (MM) pharmacokinetic analysis using the NONlinear Mixed Effects Modeling (NONMEM) software. Within the broader thesis investigating advanced parameter estimation for saturable elimination processes, this guide provides the foundational application notes and protocols. Accurate estimation of Vmax (maximum elimination rate) and Km (substrate concentration at half Vmax) is critical for predicting nonlinear pharmacokinetics, informing dosing regimens, and understanding drug-drug interactions [10]. NONMEM, the industry-standard tool for population analysis, is particularly suited for this task as it employs nonlinear mixed-effects modeling to simultaneously analyze sparse or heterogeneous data from all individuals, providing superior accuracy and precision compared to traditional linearization methods [19] [10].
Prerequisites: Data Structure and Software Environment
Successful MM analysis in NONMEM requires correctly formatted data and an appropriate software ecosystem.
2.1 Data File Structure and Requirements The data file is a comma- or space-delimited text file where each row represents a record (dosing or observation) for an individual [19]. For MM analysis, specific data items are mandatory. The core structure is outlined below:
Table 1: Essential Data Items for NONMEM Analysis using PREDPP
| DATA ITEM | DESCRIPTION | REQUIREMENT for MM Analysis | EXAMPLE |
|---|---|---|---|
ID |
Unique subject identifier | Mandatory | 1, 2, 3 |
TIME |
Elapsed time since start of analysis | Mandatory | 0, 2.5, 5.0 |
AMT |
Dose amount (for dosing records) | Mandatory for dosing events | 1000, 0 |
CMT |
Compartment number | Required if using ADVAN subroutines | 1, 2 |
EVID |
Event identifier (0=obs., 1=dose, etc.) | Mandatory | 0, 1, 4 |
DV |
Dependent variable (observed concentration) | Mandatory for observation records | 45.2, 12.7 |
MDV |
Missing dependent variable indicator | Recommended (1=missing, 0=present) | 0, 1 |
RATE |
Infusion rate (if applicable) | Optional for zero-order inputs | -2 (for modeled rate) |
For a typical single-compartment model with Michaelis-Menten elimination (ADVAN10), the central compartment is designated as CMT=1 and the output compartment as CMT=2 [19]. Accurate EVID and MDV coding is crucial for NONMEM to correctly process the data stream [19].
2.2 Software and System Requirements NONMEM is written in ANSI Fortran 95 and requires a compatible compiler (e.g., Intel Fortran, gFortran) [20]. As model estimation can be computationally intensive, a fast machine with at least 1-2 GB of dedicated memory is recommended [20]. The software is licensed annually through ICON plc [20]. Effective analysis is facilitated by a suite of supporting tools:
Table 2: Supporting Software Ecosystem for NONMEM Workflow
| Tool Category | Example Software | Primary Function |
|---|---|---|
| Graphical User Interface (GUI) | Pirana, Census 2 [34] | Provides an environment to run, manage, and edit models, and interpret output. |
| Scripting & Automation | Perl Speaks NONMEM (PsN) [19] [34] | Aids in model development through tools for validation (bootstrap, VPC), covariate screening, and data handling. |
| Statistical & Graphical Analysis | R (with packages like xpose, IQRtools) [35] [34] |
Used for data preparation, exploratory analysis, generation of diagnostic plots, and custom post-processing. |
| Code Editor | Notepad++ (with NONMEM syntax highlighting) [35] | Facilitates writing and debugging control stream files. |
Core Experimental Protocol for MM Analysis
This protocol outlines the steps to implement and estimate a Michaelis-Menten model in NONMEM, based on best practices and simulation studies [10].
3.1 Control Stream Configuration for MM Kinetics
The control stream (filename.ctl) directs all modeling actions. Key code blocks for a basic MM model are:
$PROB: Define the problem title.$INPUT: List the data items (ID, TIME, AMT, DV, CMT, EVID, ...) in the exact order they appear in the data file.$DATA: Specify the data file name and options (e.g.,IGNORE=@to ignore header lines).$SUBROUTINE: SelectADVAN10for a one-compartment model with Michaelis-Menten elimination andTRANS1[19]. This directly provides the parameters VM (Vmax) and KM (Km).$PK: Define the population (typical) parameters and inter-individual variability (IIV).
$ERROR: Define the residual error model. For MM analysis, a combined additive and proportional error model is often appropriate [10]:
$THETA,$OMEGA,$SIGMA: Provide initial estimates for fixed effects (THETAs), variances of IIV (OMEGAs), and residual error (SIGMA).$ESTIMATION: Choose the estimation method. For MM models, First Order Conditional Estimation with interaction (FOCEI) or the more advanced Stochastic Approximation Expectation-Maximization (SAEM) are recommended [32] [10].
3.2 Workflow Diagram: From Data to Validated Model The following diagram illustrates the iterative, cyclical workflow for population PK/PD model development in NONMEM, from data preparation to final model validation [35].
Diagram: Iterative Workflow for Population PK/PD Model Development
3.3 Selection of Estimation Method The choice of estimation algorithm is critical for accurate MM parameter estimation. A simulation study demonstrated that nonlinear methods in NONMEM outperform traditional linearization techniques (Lineweaver-Burk, Eadie-Hofstee) [10].
Table 3: Comparison of Estimation Methods for MM Parameters [32] [10]
| Method | Description | Key Characteristic | Suitability for MM |
|---|---|---|---|
| First Order (FO) | Linearizes inter- and intra-individual variability. | Fast but approximate; biased with high variability/sparse data. | Low - Not recommended for final estimation. |
| FOCE with Interaction | Conditional estimation linearizing residual error. | More accurate than FO for most problems. | High - Standard reliable choice. |
| Laplace | Similar to FOCE but uses second-order approximation. | Used for highly nonlinear models or non-normal data. | Medium - Can be tried if FOCE fails. |
| Stochastic Approximation EM (SAEM) | Monte Carlo method using Markov Chain sampling. | Accurate for complex models; handles sparse data well. | Very High - Often optimal for MM kinetics [32] [10]. |
| Importance Sampling (IMP) | Monte Carlo EM method sampling around the mode. | Provides precise objective function value. | High - Good for final evaluation after SAEM. |
| Iterative Two Stage (ITS) | Approximate EM method using conditional modes. | Faster than FOCE but less accurate for sparse data. | Medium - Useful for initial exploratory runs. |
Based on the comparative study, the SAEM method (available in NONMEM 7.6) followed by a final IMP evaluation is highly recommended for MM analysis as it provides the most accurate and precise estimates, especially with realistic combined error structures [32] [10].
3.4 Model Components and Parameter Relationships A clear understanding of the model components and their mathematical relationships is fundamental. The following diagram depicts the structure of a one-compartment model with Michaelis-Menten elimination and the flow of parameters through the modeling process.
Diagram: Components and Relationships in a Michaelis-Menten PK Model
Validation and Diagnostic Protocol
4.1 Standard Model Evaluation Metrics A robust MM model must pass several diagnostic checks [35]:
- Successful Estimation: Minimum of the objective function found, with successful covariance step.
- Parameter Precision: Relative standard errors (RSE%) for fixed and random effects typically <30-50%.
- Diagnostic Plots: Observations vs. Population (PRED) and Individual (IPRED) predictions should be scattered evenly around the line of identity. Conditional weighted residuals (CWRES) vs. PRED or TIME should be randomly scattered around zero.
- Visual Predictive Check (VPC): A simulation-based check where the majority of observed data falls within the prediction intervals of simulated data, confirming the model adequately describes central tendency and variability.
4.2 Simulation-Based Validation (Bootstrap) Internal validation is performed using the nonparametric bootstrap [35]:
- Resample: Generate 500-1000 bootstrap datasets by randomly sampling subjects with replacement from the original dataset.
- Refit: Re-estimate the final model on each bootstrap dataset.
- Analyze: Calculate the median and 95% confidence interval of the parameter estimates from all successful runs. Compare these intervals with the original parameter estimates; close agreement indicates model stability.
4.3 Quantitative Validation from Comparative Studies A simulation study provides quantitative benchmarks for expected performance. When estimating MM parameters from in vitro-like elimination kinetic data, NONMEM's nonlinear methods (NM in the study) showed superior accuracy and precision [10].
Table 4: Validation Metrics from Simulation Study [10]
| Estimation Method | Accuracy (Median Bias) | Precision (90% CI Width) | Key Finding |
|---|---|---|---|
| NONMEM (NM) | Lowest bias | Narrowest CI | Most reliable and accurate method. |
| Traditional Linearization (LB, EH) | Higher bias | Wider CI | Performance worsens with combined error models. |
| Other Nonlinear Regression (NL, ND) | Moderate bias | Moderate CI | Better than linearization but inferior to NM. |
This evidence strongly supports the use of NONMEM's full time-course nonlinear estimation over methods relying on initial velocity calculations or linear transformations for MM analysis [10].
The Scientist's Toolkit for MM Analysis
Table 5: Essential Research Reagent Solutions and Materials
| Tool/Resource | Category | Function/Explanation |
|---|---|---|
| NONMEM 7.6+ | Core Software | Industry-standard engine for nonlinear mixed-effects modeling. Essential for population MM parameter estimation [20] [32]. |
| Pirana | Workflow GUI | Manages run execution, output comparison, and integrates with PsN/R. Critical for organizing complex modeling projects [34]. |
| Perl Speaks NONMEM (PsN) | Automation Toolkit | Executes essential validation techniques like bootstrap and VPC. Required for rigorous model qualification [35] [34]. |
R with xpose/ggplot2 |
Diagnostic Graphics | Creates standardized diagnostic plots (e.g., OV vs. PRED, CWRES) for model evaluation and publication [35]. |
| Simulated MM Datasets | Validation Reagent | Used for method qualification and troubleshooting. The study by Cho et al. (2018) provides an excellent template for generating validation data [10]. |
| SAEM Estimation Method | Algorithm | The preferred estimation method in NONMEM for MM models due to its accuracy in handling nonlinear kinetics and combined error structures [32] [10]. |
| Fractional Differential Equation Subroutine | Advanced Tool | For modeling anomalous kinetics (power-law behavior). Useful for extending MM analysis to complex systems where standard ODEs fail [17]. |
| Nonparametric Estimation (NONP) | Diagnostic Tool | Evaluates the shape of parameter distributions. Helps diagnose and correct for bias when inter-individual variability is non-normal [26]. |
Implementing the Michaelis-Menten Model in NONMEM: Code, Estimation, and Application
Within the specialized field of pharmacometrics, the accurate estimation of Michaelis-Menten (MM) parameters (VM and KM) is critical for characterizing the nonlinear, saturable elimination kinetics exhibited by numerous therapeutic agents. This application note is framed within a broader thesis investigating advanced methodologies for MM parameter estimation using NONMEM (NONlinear Mixed Effects Modeling), the industry-standard software for population pharmacokinetic/pharmacodynamic (PK/PD) analysis [19]. While NONMEM's PREDPP library provides a dedicated, analytically solved one-compartment MM model (ADVAN10), complex real-world scenarios—such as multi-compartment distribution with MM elimination or intricate absorption models—necessitate a more flexible approach [36] [37]. This document details the implementation of these complex MM structures using PREDPP's general differential equation solvers (ADVAN6, ADVAN8, ADVAN13, etc.) and the $DES subroutine, bridging a gap between standard functionality and advanced research needs [38] [39]. The protocols herein are designed for researchers and drug development professionals requiring robust, customizable models for precise parameter estimation.
Core Components: PREDPP, ADVAN, and $DES
The NONMEM system consists of three primary components: the estimation engine (NONMEM), the translator (NM-TRAN), and the pharmacokinetic prediction suite (PREDPP) [18]. PREDPP contains a library of ADVAN (ADVANCE) subroutines, each representing a specific PK model or class of models [40] [41]. The user selects an ADVAN via the $SUBROUTINES record in the control stream.
Table 1: Key PREDPP ADVAN Subroutines for Linear and Michaelis-Menten Kinetics
| ADVAN | Model Description | Solution Type | Basic Parameters (TRANS1) | Typical Use Case |
|---|---|---|---|---|
| ADVAN1 | One-compartment linear | Analytical | K, V | Simple IV bolus kinetics [40] [19] |
| ADVAN2 | One-compartment with 1st-order absorption | Analytical | KA, K | Oral dosing [40] [19] |
| ADVAN3 | Two-compartment linear | Analytical | K, K12, K21 | IV bolus with distribution [40] [19] |
| ADVAN10 | One-compartment with Michaelis-Menten elimination | Analytical | VM, KM | Standard saturable elimination [40] [37] |
| ADVAN5/7 | General linear model (up to 999 comps) | Matrix Exponential | User-defined | Complex linear compartmental structures [36] [38] |
| ADVAN6/8/13 | General nonlinear model | ODE Solution | User-defined | Custom MM models, complex kinetics [38] [37] |
For models beyond the predefined set (ADVAN1-4, 10-12), PREDPP offers general subroutines. ADVAN6, ADVAN8, and ADVAN13 are used for models defined by ordinary differential equations (ODEs) [38] [37]. When using these, the user must provide a $DES block in the control stream (or a separate DES Fortran subroutine). This block contains the code that defines the right-hand side of the differential equations governing drug movement between compartments [39]. This is the essential mechanism for coding custom MM elimination within a multi-compartment structure or alongside other nonlinear processes.
Workflow for Selecting ADVAN Subroutines for Michaelis-Menten Models
Methodology: Coding MM Elimination with $DES
The implementation of a user-defined MM model involves a sequential configuration of NM-TRAN records. The following protocol outlines the steps for coding a two-compartment model with Michaelis-Menten elimination from the central compartment.
Step 1: Define the Model Structure with $MODEL
The $MODEL block declares the number and type of compartments. For a two-compartment mammillary model with a peripheral tissue compartment and an optional output compartment, the code is:
Step 2: Select a General Nonlinear ADVAN
The $SUBROUTINES record specifies the ODE solver. ADVAN13 (using LSODA) is often preferred for its ability to handle both stiff and non-stiff equations efficiently [37].
The TOL value controls the numerical integration precision [38].
Step 3: Declare and Assign Parameters in $PK
The $PK block is used to define the model's parameters, including fixed effects, random effects (ETAs), and covariate relationships. Typical parameters for this model include central volume (V1), inter-compartmental clearance (Q), peripheral volume (V2), and the MM parameters (VM, KM).
Step 4: Code the Differential Equations in $DES
This is the core of the custom model. The $DES block calculates the derivative (DADT) for each compartment amount (A). For the two-compartment MM model:
The equation MM_RATE implements the standard Michaelis-Menten velocity equation, where the rate of elimination is a function of the amount A(1) in the central compartment [17] [39].
Step 5: Define the Observation Model in $ERROR
Finally, the $ERROR block links the model predictions to the observed data, typically defining a residual error model.
Two-Compartment Model with Michaelis-Menten Elimination Pathway
Application Protocols and Parameter Estimation
Protocol for a Complex Absorption MM Model
Chatelut et al. (1999) modeled the absorption of alpha interferon using simultaneous first-order and zero-order processes into a one-compartment body with linear elimination [38]. Adapting this for MM elimination demonstrates the flexibility of the $DES approach.
Experimental Methodology & NM-TRAN Implementation:
- Model Schematic: Two parallel depot compartments (DEPOT1 for first-order, DEPOT2 for zero-order absorption) feed into a central compartment with MM elimination.
- $MODEL: Define three compartments:
DEPOT1,DEPOT2,CENTRAL. - $PK: Declare parameters:
KA(first-order absorption rate),D2(zero-order absorption duration),FZ(fraction absorbed via first-order),VM,KM,V. - $DES Code:
- Data File: Requires two dosing records per dose: one to
CMT=1(amount = FZDOSE) and one toCMT=2(amount = (1-FZ)DOSE).
Protocol for MM Parameter Estimation Methods
Accurate estimation of VM and KM is computationally challenging due to the nonlinearity. The choice of estimation algorithm in NONMEM is critical.
Table 2: Comparison of NONMEM Estimation Methods for Michaelis-Menten Models
| Method | Principle | Advantages for MM | Disadvantages for MM | Recommended Use |
|---|---|---|---|---|
| First Order (FO) | Linearizes random effects around zero [32]. | Fastest computation. | High bias with large inter-individual variability or sparse data [32]. | Initial model exploration only. |
| First Order Conditional Estimation (FOCE) | Linearizes around conditional estimates of ETAs [32]. | More accurate than FO, standard for nonlinear PK. | May fail with high residual error or severe nonlinearity. | Standard choice for most MM models with rich data. |
| FOCE with INTERACTION | Accounts for interaction between inter- and intra-individual error [32]. | Improved accuracy when residual error is large. | Slightly slower than FOCE. | Default choice for final MM model estimation. |
| Importance Sampling (IMP) / SAEM | Monte Carlo methods for exact likelihood evaluation [32]. | Most accurate for complex nonlinearities and sparse data. | Computationally intensive, stochastic variability in estimates. | Complex models, problematic convergence with FOCE. |
Recommended Estimation Protocol:
- Initialization: Use literature or naive pooled estimates for
THETAinitial values. SetOMEGAdiagonal elements to moderate values (e.g., 0.09 for 30% CV). - Step 1 - Preliminary Exploration: Run FO estimation to check data structure and identify obvious problems.
- Step 2 - Base Model Estimation: Switch to FOCE with INTERACTION. Ensure successful termination and examine gradient messages.
- Step 3 - Covariate Modeling: Continue with FOCE-INTERACTION to test significant covariates on parameters like
VM. - Step 4 - Final Validation: If convergence is difficult or data are sparse, confirm final parameter estimates using a Monte Carlo method (IMP or SAEM) [32].
Advanced Applications and the Scientist's Toolkit
Incorporating Fractional Differential Equations (FDEs)
Recent research extends nonlinear mixed-effects modeling to systems defined by Fractional Differential Equations (FDEs), which describe anomalous kinetics and memory effects [17]. A one-compartment model with MM elimination can be reformulated as a fractional FDE:
where ^C_0D^α_t is the Caputo fractional derivative of order α (0<α<1) [17]. Implementing this in NONMEM requires a user-supplied subroutine that replaces the $DES block with a numerical solver for FDEs, demonstrating the extensibility of the platform for cutting-edge MM research [17].
The Scientist's Toolkit: Essential Components
Table 3: Research Reagent Solutions for MM Modeling with $DES
| Tool/Component | Function in MM Model Development | Key Considerations |
|---|---|---|
| PREDPP Library (ADVAN10) | Provides a pre-built, analytically solved one-compartment MM model for validation and comparison [40] [37]. | Serves as a benchmark for custom $DES models. |
| General Nonlinear ADVAN (6,8,13) | Provides the ODE solver framework necessary for implementing custom MM structures [38] [37]. | Choice depends on model stiffness; ADVAN13 (LSODA) is often robust. |
$DES Block |
The core "reagent" for coding the differential equations that define the custom MM kinetic process [39]. | Must correctly calculate derivatives DADT. Use compact array format for large models. |
| TOL Option | Sets the tolerance for the numerical ODE solver, controlling precision and stability [38] [37]. | A value of 4-6 is typical. Adjust if runs fail due to integration error. |
| MU Referencing | A coding technique (using MU_ prefix) that improves estimation efficiency, especially with EM methods [32]. |
Highly recommended for complex models to speed up convergence. |
| PDx-Pop, Pirana, PsN | Third-party modeling environments that provide workflow management, visualization, and automated tools (e.g., VPC, bootstrap) [19]. | Essential for efficient, reproducible, and robust model development. |
Implementing Michaelis-Menten pharmacokinetics using the $DES subroutine and general ADVANs in PREDPP unlocks a powerful paradigm for pharmacometric research. This approach transcends the limitations of precompiled solutions, allowing researchers to construct and estimate parameters for sophisticated, mechanism-based models that accurately reflect complex biological processes—from multi-compartment saturation kinetics to parallel absorption pathways. Mastery of this technique, combined with a strategic understanding of estimation algorithms and diagnostic tools, positions scientists to tackle challenging nonlinear PK problems, ultimately contributing to more informed drug development decisions and optimized therapeutic regimens. The methodologies outlined here provide a foundational protocol that can be adapted and extended as part of an advanced thesis in modern pharmacometric analysis.
Within the broader thesis on advanced pharmacometric modeling using NONMEM, the generation of robust initial estimates for Michaelis-Menten parameters—the maximum elimination rate (Vmax) and the substrate concentration at half Vmax (Km)—is not a mere preliminary step but a critical determinant of research success. Nonlinear mixed-effects models, which are fundamental to population pharmacokinetic (PK) analysis, are inherently dependent on adequate initial parameter estimates for efficient and correct convergence of the estimation algorithm [42]. Poor initial estimates can lead to failed minimization, termination at local minima yielding biologically implausible parameter values, or protracted model-building cycles plagued by instability [43].
This challenge is particularly acute for Michaelis-Menten kinetics, a cornerstone model for characterizing saturable enzymatic elimination. The parameters Vmax and Km are often correlated, and their estimation can be sensitive to the design and quality of the data [13]. Framed within a thesis exploring NONMEM's full potential, this document provides detailed application notes and protocols. It moves beyond simplistic rule-of-thumb guesses, presenting a systematic, multi-strategy toolkit for generating scientifically defensible initial estimates. These strategies are designed to enhance model reliability, reduce development time, and form the foundation for robust covariate analysis and clinical simulations in model-informed drug development [43] [44].
Computational Foundations: NONMEM and Michaelis-Menten Kinetics
2.1 NONMEM Architecture for PK Modeling
NONMEM (NONlinear Mixed Effects Modeling) is a software program that fits PK/PD models to data while accounting for inter-individual and intra-individual variability [19]. Its workflow involves a control stream (.ctl file), a data file, and output files. The control stream specifies the model, parameters, and estimation methods. A key component is PREDPP, a library of pre-compiled PK models accessed via the $SUBROUTINES record using ADVAN and TRANS subroutines [19].
2.2 Implementing Michaelis-Menten Elimination in NONMEM
For Michaelis-Menten kinetics, the ADVAN10 subroutine is used in conjunction with TRANS1. This specifies a one-compartment model with Michaelis-Menten elimination from the central compartment. The two basic parameters defined in the $PK block are VM (Vmax) and KM (Km) [19]. The structural model is defined by the differential equation: dA/dt = - (VM * A/V) / (KM + A/V), where A is the amount in the central compartment and V is the volume of distribution. Proper data setup is crucial: the dataset must include ID, TIME, AMT (dose), DV (observed concentration), EVID (event identifier), and MDV (missing dependent variable) items, with records sorted by ID and TIME [45].
2.3 The Imperative for Robust Initial Estimates The estimation process in NONMEM is an iterative search for parameter values that minimize an objective function value (OFV). The landscape of this search can be complex, with multiple local minima. Initial estimates that are far from the true values can cause the algorithm to fail to converge or to converge to an incorrect solution [43] [46]. This is formally linked to the balance between model complexity and the information content of the data; an over-parameterized model relative to the data will exhibit instability, for which poor initial estimates are a primary catalyst [43]. Therefore, a robust initialization strategy directly addresses the root cause of a common class of model instability issues.
Strategic Framework for Generating Initial Estimates
A multi-faceted approach is recommended, moving from simple, data-informed guesses to sophisticated, automated procedures. The choice of strategy depends on data type (rich vs. sparse, in vitro vs. in vivo), available prior knowledge, and the stage of analysis.
3.1 Strategy 1: Prior Knowledge and Literature-Based Anchoring The first and most straightforward strategy involves leveraging existing knowledge.
- Published Values: For a known enzyme or transporter, literature values for Km and Vmax (often scaled to per mg protein or per cell) can provide an order-of-magnitude starting point. Allometric scaling (e.g., using body weight exponents of 0.75 for clearance) may be applied to extrapolate in vitro Vmax to an in vivo whole-organ value [44].
- Related Compounds: Data for structurally or mechanistically similar compounds can offer plausible ranges.
- Biological Plausibility: Estimates should be checked for biological sense (e.g., Km should be within or near the range of observed substrate concentrations).
3.2 Strategy 2: Data-Driven Methods from In Vitro and Rich Data For in vitro enzyme kinetic studies or rich in vivo PK data, direct graphical or computational analysis can yield excellent initial estimates.
- Non-Compartmental Analysis (NCA): For in vivo data following a single dose, NCA can estimate total clearance (CL) and the area under the curve (AUC). At low, non-saturating concentrations, clearance is approximated by Vmax/Km. A rough estimate can be obtained:
Km ≈ Dose / (2 * Vd), andVmax ≈ CL * Km, where Vd is the volume of distribution from NCA [42]. - Visual Inspection of Saturation: Plotting elimination rate (or observed concentration-time slope) against concentration can visually indicate the range of Km (concentration where the rate is at half-maximum) and Vmax (the plateau rate).
- Linearized Plots (For In Vitro Data Only): While final estimation should use nonlinear methods, linear transformations like the Lineweaver-Burk (1/v vs. 1/[S]) or Eadie-Hofstee (v vs. v/[S]) plot can provide quick, albeit statistically biased, initial guesses from in vitro velocity data [13].
3.3 Strategy 3: Automated and Preconditioning Pipelines For population modeling with sparse data or during automated model development, algorithmic generation of initial estimates is essential.
- Automated Base Model Pipelines: Recent research has developed integrated pipelines that use data-driven algorithms to compute initial estimates for both structural and statistical parameters. These pipelines perform well with both rich and sparse data scenarios, generating estimates that lead to final parameters closely aligned with true or literature values [42].
- Preconditioning with Simplified Models: A highly effective strategy is to "precondition" the problem by first fitting a simpler, more stable model.
- Fit a linear elimination model (e.g.,
ADVAN1orADVAN2) to obtain estimates for clearance (CL) and volume (V). - Fix the volume parameter (V) from the linear model. Use the final CL estimate as an initial estimate for the ratio
Vmax/Km. - Provide an initial Km estimate that is within the range of observed concentrations (e.g., the median observed concentration).
- Calculate the corresponding Vmax initial estimate as
CL * Km. - Fit the full Michaelis-Menten model (
ADVAN10), freeing all parameters. This sequential approach often leads to more stable convergence than starting with naïve guesses.
- Fit a linear elimination model (e.g.,
Table 1: Comparison of Estimation Method Performance for Michaelis-Menten Parameters (Simulation Study) [13]
| Estimation Method | Description | Key Advantage | Key Limitation | Relative Performance (Accuracy & Precision) |
|---|---|---|---|---|
| Lineweaver-Burk (LB) | Linear fit to 1/v vs. 1/[S] plot. | Simple, visual. | Prone to statistical bias; poor error model assumption. | Least accurate, especially with high variability. |
| Eadie-Hofstee (EH) | Linear fit to v vs. v/[S] plot. | Less bias than LB. | Still assumes erroneous error structure. | Low to moderate accuracy. |
| Nonlinear Regression (NL) | Direct nonlinear fit of v to [S] data. | Correct error structure. | Requires initial velocity data, discards time-course info. | High accuracy. |
| Nonlinear Dynamic (NM) | Nonlinear fit of full [S]-time course data (e.g., via NONMEM). | Uses all data; correct error model; estimates in vitro half-life. | Requires appropriate software; needs careful initialization. | Most accurate and precise, superior with combined error models. |
Detailed Experimental Protocols
4.1 Protocol 1: Generating and Analyzing In Vitro Kinetic Data for Initial Estimates
- Objective: To obtain initial estimates for Vmax and Km from an in vitro drug elimination experiment using primary hepatocytes or microsomes.
- Materials: See "The Scientist's Toolkit" below.
- Workflow:
- Incubation: Incubate the test compound at 5-8 different substrate concentrations (spanning expected Km) with the enzyme system. Use a short, linear time course (e.g., 5-7 time points per concentration).
- Data Generation: Quench reactions and quantify substrate depletion or metabolite formation. Calculate the initial velocity (v) for each [S] using linear regression of amount vs. time for the early, linear phase.
- Initial Estimate Calculation:
- Method A (Linearization): Create an Eadie-Hofstee plot (v vs. v/[S]). Perform a linear regression. The y-intercept is Vmax. The slope is -Km.
- Method B (Direct Plot): Plot v vs. [S]. Visually estimate Vmax (plateau) and Km (concentration at half Vmax).
- NONMEM Preparation: Format a dataset with columns:
ID(incubation ID),TIME,CONC([S]),DV(measured concentration),EVID,MDV. UseEVID=0for observations. - Control Stream Setup: Use
$SUBROUTINES ADVAN10 TRANS1. In$PK, declareVM = THETA(1) * EXP(ETA(1))andKM = THETA(2) * EXP(ETA(2)). Input the graphical estimates as initial values forTHETA(1)andTHETA(2).
- Validation: The final NONMEM estimates from the full time-course analysis (NM method) should be more reliable than the graphical initials [13].
4.2 Protocol 2: Preconditioning Strategy for In Vivo Population PK Analysis
- Objective: To generate robust initial estimates for a population Michaelis-Menten model using clinical PK data.
- Workflow Diagram:
- Protocol Steps:
- Data Preparation: Ensure the dataset is correctly formatted for PREDPP: sorted by
IDandTIME, with properEVIDandMDVflags [45]. - Linear Model Fit:
- Build a one- or two-compartment linear model (
ADVAN1/ADVAN2orADVAN3/ADVAN4). - Use reasonable initials (e.g., from NCA or literature).
- Run the model to successful convergence. Document the final estimates for clearance (
CL) and central volume (V).
- Build a one- or two-compartment linear model (
- Calculate Michaelis-Menten Initials:
- Set
Km_initialto the median of the observed concentrations (DVwhereEVID=0). - Calculate
Vmax_initial = CL_final * Km_initial.
- Set
- Build and Run Michaelis-Menten Model:
- Create a new control stream using
$SUBROUTINES ADVAN10 TRANS1. - In the
$PKblock, define: - On the
$THETArecord, set:(Vmax_initial, , ) (Km_initial, , ) (V_final, FIXED). - Run the model. The successful convergence of this preconditioned model provides the final robust parameter estimates.
- Create a new control stream using
- Data Preparation: Ensure the dataset is correctly formatted for PREDPP: sorted by
Table 2: Key Diagnostic Outputs from NONMEM for Validating Model Convergence [46]
| Output File/Item | Description | Interpretation for Successful Convergence |
|---|---|---|
| Report (.lst/.res) File | Primary results file. | Check for MINIMIZATION SUCCESSFUL and absence of severe warnings. |
| Objective Function Value (OFV) | -2 log-likelihood. | Should stabilize at final iteration. Large changes indicate problems. |
| Parameter Estimates (THETA) | Final population parameter values. | Should be biologically plausible and have reasonable precision. |
| Standard Errors | Precision of THETA estimates. | Relative standard error (RSE) < 30-50% often desired. Failed covariance step may indicate instability. |
| Gradients | Derivative of OFV w.r.t parameters at solution. | All gradients should be near zero (e.g., < 0.001). |
| .ext File | Final estimates table. | Contains termination codes, OFV, and parameter estimates for easy extraction. |
| .phi File | Individual parameter estimates (ETA). | Useful for diagnosing individual fit problems and visualizing IIV. |
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions and Computational Tools
| Item | Function in Initial Estimate Generation |
|---|---|
| Graphical/Analysis Software (R, Python) | For data preprocessing, NCA, creating diagnostic plots (Eadie-Hofstee), and running automated estimation pipelines [42] [13]. |
| NONMEM Software | Industry-standard platform for nonlinear mixed-effects modeling. The ADVAN10 subroutine is specifically designed for Michaelis-Menten kinetics [19]. |
| PKNCA R Package | Performs Non-Compartmental Analysis to obtain CL and AUC, which inform the Vmax/Km ratio [42]. |
| Automated Pipeline Scripts | Custom or published R scripts that algorithmically derive initial estimates for structural and statistical parameters from the dataset, reducing manual guesswork [42]. |
| Diagnostic Toolkit (Xpose, Pirana) | Assists in evaluating model convergence, goodness-of-fit, and comparing OFV between linear and nonlinear models, confirming the need for Michaelis-Menten kinetics. |
| Estimation Algorithms (FOCE, SAEM) | Within NONMEM, the First Order Conditional Estimation (FOCE) and Stochastic Approximation EM (SAEM) methods are robust algorithms for fitting nonlinear models like Michaelis-Menten [13]. |
Diagram: NONMEM Michaelis-Menten Model Specification & Estimation Workflow
Within the context of a broader thesis on Michaelis-Menten parameter estimation research using NONMEM, the selection of an appropriate estimation algorithm is not merely a technical step but a foundational scientific decision. Nonlinear mixed-effects (NLME) modeling in pharmacometrics aims to identify the population parameters (THETAs, OMEGAs, SIGMAs) that best describe the observed data while accounting for inter-individual (ETA) and residual (EPS) variability [32]. The core computational challenge lies in evaluating the integral over all possible individual parameter values for each subject [32]. Different estimation methods solve this problem with distinct algorithmic strategies, leading to significant differences in performance, reliability, and efficiency [47] [48] [49].
For researchers focused on enzyme kinetics, particularly the estimation of V~max~ and K~m~ via the Michaelis-Menten equation, this choice is critical. While traditional linearization methods (e.g., Lineweaver-Burk) are available, nonlinear estimation methods within NONMEM provide more reliable and accurate parameter estimates, especially when data incorporates complex error structures [24]. This article provides a detailed comparison of four key estimation families in NONMEM: First-Order Conditional Estimation (FOCE), Importance Sampling Expectation-Maximization (IMP), Stochastic Approximation Expectation-Maximization (SAEM), and Bayesian (BAYES) approaches. It offers application notes, experimental protocols, and practical guidance to inform method selection within advanced pharmacometric research.
Characteristics and Algorithmic Foundations of Estimation Methods
2.1 First-Order Conditional Estimation (FOCE)
FOCE is a classical maximum-likelihood method that approximates the likelihood integral. It evaluates the mode (most likely values) of the individual parameters (ETAs) and uses a first-order (linear) approximation of the variance around this mode to estimate the integral [32]. The INTERACTION option (often termed FOCE-I) accounts for correlation between inter-individual and residual error and is recommended when residual error is proportional [49]. While faster than exact methods for simpler problems, its approximation can lead to inaccuracies or convergence failures with complex models, high inter-individual variability, or sparse data [48] [32]. The FAST option can significantly reduce run time by leveraging a specific model structure [48] [49].
2.2 Expectation-Maximization (EM) Methods: IMP and SAEM EM methods are "exact" algorithms that perform a Monte Carlo integration to explore the entire ETA space [32]. They consist of an Expectation (E) step, which evaluates the integral, and a Maximization (M) step, which updates the population parameters.
- Importance Sampling (IMP): The E-step uses a normal sampler centered at the mode (or mean) of the conditional density, focusing on the "important" region of the parameter space. It provides accurate objective function values and standard errors directly [32].
- Stochastic Approximation EM (SAEM): This method uses a Markov Chain Monte Carlo (MCMC) sampler. It often runs in two phases: a first phase with rapid parameter updates and a second phase where individual parameter samples are averaged for convergence. SAEM itself does not evaluate the objective function; a final
IMP EONLY=1step is typically used to obtain it [47] [32].
2.3 Bayesian (BAYES) Approach The Bayesian method in NONMEM employs MCMC sampling to characterize the full posterior distribution of the parameters, given the data and prior information [20] [50]. It is particularly useful for complex models where maximum likelihood methods struggle, for incorporating prior knowledge, and for directly quantifying parameter uncertainty. Unlike EM methods that seek a single "most likely" estimate, BAYES generates a large sample of probable parameter sets [50].
2.4 Software and System Context
All these methods are implemented in NONMEM, the industry-standard software for population PK/PD analysis [20]. The software comprises the NONMEM engine, the PREDPP subroutine library for PK models, and the NM-TRAN preprocessor [20]. The choice of estimation method interacts with the model definition, particularly the selection of $SUBROUTINE (e.g., analytical ADVAN routines vs. differential equation solver ADVAN13), which dictates whether the problem is solved analytically or via numerical integration [49].
Table 1: Core Characteristics of NONMEM Estimation Methods [47] [48] [32]
| Method | Algorithmic Class | Key Algorithmic Feature | Computational Demand | Typical Convergence Behavior | Objective Function (OFV) |
|---|---|---|---|---|---|
| FOCE(-I) | Classical MLE | First-order approximation around ETA mode. | Low to Moderate for simple models; increases with complexity. | Can fail with complex models, sparse data, or high IIV. | Directly estimated, used for hypothesis testing (LRT). |
| IMP | Exact Monte Carlo EM | Importance sampling from a normal proposal density. | High (requires many samples per subject). | Robust for complex models; sensitive to model misspecification [48]. | Directly estimated, used for hypothesis testing (LRT). |
| SAEM | Exact Monte Carlo EM | MCMC sampling with stochastic approximation. | Moderate to High. | Robust; benefits from ITS or parameter log-transform for difficult problems [47]. |
Not directly estimated; requires a final IMP EONLY step. |
| BAYES | Bayesian MCMC | Full posterior sampling via MCMC (NUTS). | Very High. | Robust for complex models; requires careful chain diagnostics. | Not applicable; inference based on posterior distributions. |
Performance Comparison and Selection Guidelines
3.1 Empirical Findings from Comparative Studies Research indicates that the optimal method is highly context-dependent. For relatively simple PK models (e.g., one- or two-compartment) with rich data, FOCE and FOCE FAST are often the most efficient, providing precise estimates with the shortest run times [48]. However, for complex models—such as target-mediated drug disposition (TMDD), parent-metabolite systems, or models with data below the quantification limit (BLQ)—EM and Bayesian methods show superior reliability.
A study on a TMDD model with limited data (15 subjects) found IMP and IMPMAP to be the most robust, achieving stable convergence where FOCE succeeded in only 21% of runs [47]. SAEM, while sometimes having initial convergence issues, produced the most accurate individual parameter estimates (posthocs) when it did converge [47]. Another study on a complex parent-metabolite model confirmed that FOCE could fail to reliably assess parameter precision, while IMP and SAEM performed well, with SAEM being faster [48].
For Michaelis-Menten parameter estimation specifically, a simulation study demonstrated that nonlinear estimation methods in NONMEM provided more accurate and precise estimates of V~max~ and K~m~ compared to traditional linearization methods, particularly under a combined error model [24].
3.2 Selection Framework and Practical Recommendations Method selection should be guided by model complexity, data structure, and project goals.
Table 2: Estimation Method Selection Guide [47] [48] [32]
| Scenario | Recommended Primary Method(s) | Rationale and Practical Notes |
|---|---|---|
| Simple PK Model (1-2 CMTs), Rich Data | FOCE-I (with FAST option) |
Fastest and most efficient. Use as benchmark. |
| Complex Model (TMDD, ODEs, >6 params) | SAEM or IMP | More reliable convergence. Start SAEM, use IMP EONLY for final OFV. |
| Sparse Data or High IIV | SAEM or IMP | Better handles poor parameter identifiability. |
| Model with BLQ Data | SAEM or IMP | More accurately handles censored data likelihood. |
| Small Sample Size (N<10) | FOCE-I or BAYES | One study found FOCE-I more reliable than BAYES for very small N with simple models [50]. |
| Including Prior Information | BAYES | Natural framework for incorporating prior knowledge from literature or previous studies. |
| Initial Model Exploration | ITS | Very fast approximate EM method for initial screening [32]. |
| Final Model Refinement & Covariate Testing | IMP | Provides a stable, exact OFV for likelihood ratio tests. |
General Workflow Advice:
- Start Simple: Use FOCE-I for initial model building.
- Switch for Complexity: If FOCE fails to converge or yields unstable covariance steps, switch to SAEM.
- Finalize with IMP: Use SAEM for estimation, followed by a final
IMP EONLY=1step withMAXEVAL=0to obtain the OFV and standard errors for final reporting and hypothesis testing [47] [32]. - Utilize Parallel Computing: For IMP or SAEM, use parallel computing (
NPROC) to significantly reduce runtime [20] [48].
Experimental Protocols for Method Comparison
4.1 Protocol 1: Simulation-Based Comparison for a Michaelis-Menten Elimination Model This protocol is designed to evaluate method performance for a core thesis model.
- Objective: To compare the bias, precision, and convergence rate of FOCE, IMP, SAEM, and BAYES in estimating V~max~ and K~m~ under varying levels of inter-individual variability (IIV) and residual error.
- Model: A one-compartment model with intravenous bolus administration and Michaelis-Menten elimination (e.g., using
ADVAN10orADVAN13with$DES) [49] [51]. Parameters: V~max~ (typical value), K~m~ (typical value), volume of distribution (V). IIV (log-normal) is assumed on V~max~ and K~m~. - Simulation Design:
- Subjects: Simulate 50 datasets each for N=15 (sparse) and N=50 (rich).
- Variability: Cross three levels of IIV (CV% = 20%, 50%, 80%) with two residual error models (additive and proportional).
- Software: Use
$SIMULATIONin NONMEM or themodelrpackage in R for data generation.
- Estimation: Fit each simulated dataset using all four methods.
- FOCE:
METHOD=CONDITIONAL INTERACTION MAXEVAL=9999 SIGL=9 - IMP:
METHOD=IMP ISAMPLE=1000 NITER=1000 - SAEM:
METHOD=SAEM NBURN=500 NITER=500followed byMETHOD=IMP ISAMPLE=1000 NITER=0 EONLY=1 - BAYES:
METHOD=BAYES NUTS
- FOCE:
- Outcome Measures:
- Convergence Rate: Percentage of successful minimizations per method/scenario.
- Bias: Median relative estimation error (REE) for each parameter:
(Estimate - True Value) / True Value. - Precision: Relative Root Mean Square Error (rRMSE).
- Runtime: Average computation time.
4.2 Protocol 2: Real-Data Application to a Parent-Metabolite PK System This protocol assesses methods in a real-world, complex modeling scenario [48].
- Objective: To evaluate the practical performance of estimation methods on a nonlinear parent-metabolite PK model with potential model misspecification.
- Data: Use rich PK data for oxfendazole and its metabolites (sulfone and sulfoxide) [48].
- Model Development:
- Build a base model for the parent drug using all methods.
- Develop a joint model linking parent and metabolites, incorporating formation and elimination pathways.
- Test the need for pre-systemic metabolism (a source of model nonlinearity).
- Estimation Strategy:
- Run FOCE-I as a baseline. If it fails or shows high RSEs, proceed.
- Run SAEM to obtain population parameter estimates.
- Run a final IMP step (
EONLY=1) on the SAEM estimates to obtain the OFV and covariance. - (Optional) Run BAYES to obtain full posterior distributions and compare with maximum likelihood estimates.
- Comparison Metrics:
- Success of convergence for the full joint model.
- Plausibility and precision (RSE%) of parameter estimates.
- Stability of the covariance matrix.
- Goodness-of-fit plots (e.g., population and individual predictions vs. observations).
Workflow and Decision Pathways
Diagram 1: Estimation Method Selection Algorithm This flowchart provides a practical decision pathway for selecting an estimation method based on model complexity and initial results, guiding researchers from problem assessment to final reporting [47] [48] [49].
Diagram 2: Michaelis-Menten Parameter Estimation Workflow This diagram outlines the sequential steps for robust Michaelis-Menten parameter estimation, highlighting the hybrid use of estimation methods (FOCE for exploration, SAEM for robustness, IMP for final inference) within a complete model qualification framework [24] [32].
The Scientist's Toolkit: Essential Software and Analysis Tools
Table 3: Essential Research Toolkit for NONMEM Analysis [20] [50] [51]
| Tool Name | Category | Primary Function | Role in Estimation Method Research |
|---|---|---|---|
| NONMEM 7.6 | Core Engine | Executes NLME model estimation. | Provides all estimation methods (FOCE, IMP, SAEM, BAYES). Essential for protocol execution. |
| PsN (Perl-speaks-NONMEM) | Workflow Toolkit | Automates model execution, bootstrapping, VPC, SSE. | Critical for running large simulation-estimation studies (Protocol 1) and model qualification. |
| Pirana | Modeling Environment | GUI for managing NONMEM runs, results, and diagnostics. | Facilitates comparison of outputs from different estimation methods and project management. |
| R / RStudio | Statistical Computing | Data preparation, result aggregation, custom graphics, statistical analysis. | Used to simulate data, parse NONMEM outputs, calculate performance metrics (bias, rRMSE), and generate comparative plots. |
| Xpose | R Library | Diagnostics for NLME models. | Generates standard goodness-of-fit plots to compare model performance across methods. |
| PMX Repository/Code Templates | Code Resource | Provides template control streams for common models. | Offers reliable starting code for models like Michaelis-Menten elimination, reducing setup errors [51]. |
The accurate characterization of saturable pharmacokinetic (PK) processes, predominantly described by Michaelis-Menten (M-M) kinetics, is a cornerstone of model-informed drug development for compounds exhibiting nonlinear clearance. Within the broader thesis on NONMEM-based M-M parameter estimation, this application note provides detailed protocols for simulating complex concentration-time profiles and predicting transitions into and out of saturable regimes. NONMEM, the industry-standard nonlinear mixed-effects modeling software [20], offers a suite of advanced features—including Bayesian estimation [52], delay differential equation solvers [53] [20], and optimal design evaluation [54]—that are essential for this task. Successful modeling in this context is critically dependent on robust initial parameter estimates to avoid convergence failure [7], careful handling of biological delays [53], and the strategic evaluation of trial designs to ensure parameter identifiability [54]. These protocols integrate contemporary methodologies, including automated estimation pipelines [7] and considerations for Large Language Model-assisted workflows [55], to provide a comprehensive framework for tackling the challenges of saturation kinetics.
Foundational Parameter Estimation Methods
The selection of an initial estimation strategy is pivotal for the subsequent successful estimation of M-M parameters (VM and KM). The following table summarizes the core methodologies, their applications, and key performance metrics based on validation studies.
Table 1: Summary of Initial Parameter Estimation Methods for Michaelis-Menten Modeling [7]
| Method Category | Primary Function | Key Equations/Outputs | Applicable Data Scenario | Reported Performance (rRMSE) |
|---|---|---|---|---|
| Adaptive Single-Point | Estimates CL, Vd, Ka from sparse single-point-per-individual data. | Vd = Dose / C₁; CL = Dose / Css_avg; Ka solved from 1-cpt equations. |
Sparse data, single-dose & steady-state. | Final estimates within ±15% of true simulated values. |
| Naïve Pooled NCA | Provides population PK parameter estimates via non-compartmental analysis on pooled data. | CL = Dose / AUC₀‑∞; Vz = CL / λz. |
Rich or pooled sparse data. | Serves as reliable initial estimates for base models. |
| Graphic Methods | Determines parameters via linear regression of specific PK phases (e.g., terminal elimination). | CL from slope of ln(C) vs. T; Vd ≈ Dose / C₀,extrap; Ka from residual slope. |
Rich data, clearly defined phases. | Effective for identifying linear elimination parameters pre-saturation. |
| Parameter Sweeping | Identifies optimal initial estimates for complex parameters (e.g., VM, KM) by simulating across a defined range. |
Tests candidate values; selects set minimizing relative Root Mean Squared Error (rRMSE). | All data types, essential for nonlinear parameters. | Directly optimizes initial guesses for M-M parameters prior to full NONMEM estimation. |
Experimental Scenarios for Saturational Transition Analysis
Simulating profiles that transition between linear and nonlinear kinetic zones requires well-defined experimental scenarios. The following table outlines standard and advanced scenarios used to stress-test M-M models and design clinical studies.
Table 2: Experimental Scenarios for Simulating Saturational Transitions
| Scenario Type | Primary Objective | Key Design Variables | Typical NONMEM ADVAN/TRANS | Critical Outputs |
|---|---|---|---|---|
| Rapid IV Bolus (Single Dose) | Characterize intrinsic VM and KM from a wide concentration range. |
High dose to ensure initial saturation. | ADVAN13/TRAN6 (General ODE) |
Profile shape, time to transition from zero-order to first-order elimination. |
| Oral Dosing (Multiple Ascending Dose) | Assess saturation in absorption and/or first-pass metabolism. | Dose levels, formulation (Ka), feeding state. | ADVAN13/TRAN6 or ADVAN2 (for 1-cpt oral) [56]. |
Cmax, AUC nonlinearity, bioavailability (F) changes with dose. |
| Continuous IV Infusion | Identify steady-state concentration (Css) at which clearance transitions. |
Infusion rate (R0), infusion duration. | ADVAN13/TRAN6. |
Css, time to reach steady-state, clearance value at Css. |
| Pulsatile Endogenous System | Model drug effect on an underlying saturable physiological process (e.g., hormone synthesis). | Baseline pulse amplitude, frequency, drug inhibitory IC₅₀ [57]. | ADVAN13 or ADVAN16 (for DDEs) [53]. |
Dampening of pulse amplitude, change in average baseline. |
| Optimal Design for Parameter Estimation | Optimize sampling schedule to precisely estimate VM and KM [54]. |
Sample times, number of samples, dose groups. | User-defined (e.g., ADVAN13). |
Fisher Information Matrix (FIM), predicted standard errors for VM and KM. |
Detailed Experimental Protocols
Protocol 1: Comprehensive Workflow for M-M Model Development in NONMEM
This protocol integrates initial estimation, model coding, and evaluation for a drug exhibiting saturable elimination.
1. Data Preparation and Initial Estimation:
- Standardize the dataset per
nlmixr2/NONMEM requirements (ID,TIME,AMT,DV,EVID,MDV) [56]. - Apply the automated estimation pipeline [7]:
- Use the Naïve Pooled NCA method to obtain initial estimates for linear clearance (
CLlinear) and volume (V). - Use parameter sweeping to generate initial estimates for
VMandKM. Define a plausible range (e.g.,VMfrom0.5*CLlinearto10*CLlinear;KMfrom0.1*Cmaxto2*Cmax). Simulate the M-M model and calculate rRMSE against observed data to select the best candidate pair.
- Use the Naïve Pooled NCA method to obtain initial estimates for linear clearance (
2. NONMEM Control Stream Development:
- Use a general differential equation solver (
$SUBROUTINES ADVAN13 TOL=6). - Code the M-M elimination in
$DES:
- Implement parameters with inter-individual variability (IIV) and covariate relationships in
$PK:
- Set initial estimates (
$THETA) based on the automated pipeline output and bounds forOMEGAandSIGMA[56].
3. Model Estimation & Evaluation:
- Begin estimation using the First Order Conditional Estimation with interaction (
METHOD=1 INTERACTION) [56]. - If convergence is unstable, switch to a Bayesian framework (
METHOD=BAYESorMETHOD=NUTS) [52] [20] to incorporate prior information (e.g., weakly informative priors onKMfrom in vitro data). - Evaluate goodness-of-fit (GOF) plots, parameter precision, and visual predictive checks (VPCs) to ensure the model captures the saturational transition.
Protocol 2: Implementing Delay Models for Saturable Precursor Turnover
Many saturable processes (e.g., production of an endogenous compound) involve significant time delays [53]. This protocol extends a turnover model with M-M synthesis.
1. Model Structure Definition:
- Define a precursor compartment (
A(2)) with zero-order synthesis (Kin) and first-order loss (Kout). - Define the observed response compartment (
A(1)) where the input is the delayed output from the precursor pool.
2. Delay Implementation (Choose one):
- Transit Compartment Approach: Insert
ntransit compartments betweenA(2)andA(1)to approximate a gamma-distributed delay. The mean transit time (MTT) =n / Ktr, whereKtris the transit rate [53]. - Delay Differential Equation (DDE) Approach (More Accurate): Use
ADVAN16[20]. In$DES, code:
3. Saturable Synthesis Implementation:
- Replace constant
Kinin the precursor equation with a M-M function:Kin = (VM * Stim) / (KM + Stim), whereStimcould be a drug concentration or physiological signal.
4. Estimation:
- Due to model complexity, use the Laplace estimation method (
METHOD=4 LAPLACE) or the Stochastic Approximation Expectation-Maximization (SAEM) method available in NONMEM 7.6 [20].
Visualizing Workflows and Model Architecture
Diagram 1: Integrated NONMEM Workflow for Saturable PK/PD Modeling (Max width: 760px)
Diagram 2: Architecture of a Saturable PK/PD Model in NONMEM (Max width: 760px)
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Software, Packages, and Model Components
| Tool/Component | Category | Primary Function in M-M Research | Reference/Origin |
|---|---|---|---|
| NONMEM 7.6 | Primary Software | Gold-standard for nonlinear mixed-effects modeling. Supports SAEM, Bayesian (NUTS), DDEs (ADVAN16/17), and optimal design ($DESIGN). | ICON plc [20] |
| Automated Initial Estimation Pipeline (R package) | Support Software | Generates robust initial estimates for CL, V, VM, KM from diverse (sparse/rich) datasets, critical for M-M model convergence. | [7] |
| PREDPP | Subroutine Library | Provides pre-compiled PK models (ADVAN), freeing the user from coding standard differential equations. | Bundled with NONMEM [20] |
| PsN Toolkit | Support Software | Facilitates model evaluation (VPC, bootstrap), covariate screening, and automation of NONMEM runs. | [55] |
| $DESIGN Feature | Software Feature | Evaluates and optimizes clinical trial designs via Fisher Information Matrix (FIM) to ensure precise estimation of VM and KM. | NONMEM [54] |
| Delay Differential Equation (DDE) Solvers (ADVAN16/ADVAN17) | Algorithm | Accurately models biological delays (e.g., enzyme synthesis, cell maturation) impacting saturable processes. | NONMEM 7.6 [53] [20] |
| Bayesian Estimation (METHOD=BAYES/NUTS) | Algorithm | Incorporates prior knowledge (e.g., in vitro KM) to stabilize estimation, essential for sparse data or complex models. | NONMEM [52] [20] |
| Surge/Pulse Function | Model Component | Models pulsatile endogenous baseline (e.g., growth hormone) upon which a drug acts on a saturable pathway. | [57] |
| Transit Compartment Model | Model Component | Approximates a distributed time delay using a series of compartments; simpler alternative to DDEs when variance is small. | [53] |
| Large Language Models (e.g., Claude 3.5 Sonnet) | AI Assistant | Assists in generating model structure diagrams, summarizing output, and creating report-ready tables from NONMEM results. | [55] |
The transition from robust parameter estimation to clinically actionable dosing regimens represents a critical frontier in model-informed drug development. This process is encapsulated by advanced dose optimization algorithms, such as OptiDose, which translate pharmacometric models into individualized dosing strategies [58]. Framed within a broader research thesis on NONMEM for Michaelis-Menten parameter estimation, this document details the application of these concepts. The Michaelis-Menten model, fundamental to characterizing saturable enzymatic elimination (V = Vmax * [S] / (Km + [S])), serves as a cornerstone for parameter estimation, where nonlinear estimation methods (NMs) in NONMEM have demonstrated superior accuracy and precision compared to traditional linearization techniques [24].
The optimization paradigm has evolved from merely identifying the maximum tolerated dose (MTD) to a more nuanced focus on the optimal biological dose (OBD) that balances efficacy and safety [59]. Regulatory initiatives like the FDA's Project Optimus emphasize the need for this reform, particularly in oncology, to ensure doses maximize therapeutic benefit while minimizing adverse effects [60]. The OptiDose algorithm addresses this need by mathematically formulating the dosing challenge as a finite-dimensional optimal control problem (OCP). It computes a dosing regimen (control, u) that minimizes a cost functional (J), measuring the deviation of the system's response from a desired therapeutic target (reference function) [58]. This approach has been enhanced to incorporate state constraints for managing efficacy and safety trade-offs, transforming a constrained OCP into an unconstrained problem solvable within the NONMEM framework [61].
Core Quantitative Findings and Data Synthesis
The reliability of any dose optimization exercise is predicated on the accuracy of the underlying parameter estimates. Comparative studies on Michaelis-Menten parameter estimation provide critical data for informing the initial modeling phase of the OptiDose workflow.
Table 1: Performance Comparison of Michaelis-Menten Parameter Estimation Methods [24]
| Estimation Method | Vmax Accuracy (Median Bias) | Vmax Precision (90% CI) | Km Accuracy (Median Bias) | Km Precision (90% CI) | Notes |
|---|---|---|---|---|---|
| Nonlinear Methods (NONMEM) | Highest | Narrowest | Highest | Narrowest | Most accurate/precise, especially with combined error models. |
| Lineweaver-Burk Plot | Low | Wide | Low | Wide | Traditional linearization; prone to error propagation. |
| Eadie-Hofstee Plot | Low | Wide | Low | Wide | Traditional linearization. |
| Hanes-Woolf Plot | Moderate | Moderate | Moderate | Moderate | Traditional linearization. |
| Direct Linear Plot | Moderate | Moderate | Moderate | Moderate | Traditional linearization. |
Furthermore, the capabilities of the NONMEM software itself, which is central to implementing OptiDose, have advanced significantly. The latest version, NONMEM 7.6, includes features essential for complex optimization tasks [20].
Table 2: Key NONMEM 7.6 Features Relevant for Dose Optimization [20]
| Feature Category | Specific Methods/Algorithms | Utility in Dose Optimization & M-M Research |
|---|---|---|
| Population Analysis | FOCE, Laplace, SAEM, IMP, BAYES (NUTS) | Robust estimation of population & individual M-M parameters (Vmax, Km) and their variability. |
| Advanced Solvers | ADVAN16 (for stiff delay differential equations) | Enables complex PKPD model specification for OptiDose. |
| Trial Design | Evaluation and optimal design algorithms | Informs efficient sampling for M-M parameter estimation and dose-ranging studies. |
| Parallel Computing | Multi-core/computer parallelization | Accelerates model estimation, simulation, and optimization runs. |
Experimental and Computational Protocols
Protocol 1: NONMEM-based Michaelis-Menten Parameter Estimation fromIn VitroData
This protocol establishes the foundational parameter estimates for saturable elimination pathways [24].
Objective: To accurately estimate Vmax and Km from in vitro drug elimination kinetic data using NONMEM's nonlinear estimation methods.
Materials:
- In vitro incubation data: Substrate concentration ([S]) vs. time profiles from hepatocyte or microsomal incubations.
- Software: NONMEM (v7.6 or later), interface (e.g., Pirana, PsN), R or Python for data preprocessing/post-processing.
Procedure:
- Data Preparation: Structure data in a NONMEM-compatible format (e.g., CSV). Required columns include: ID (experiment identifier), TIME, CONC (substrate concentration), and optionally, a rate column (dCONC/dt).
- Model Specification (Control Stream):
- Use
$PREDor$DESto define the differential equation:dA/dt = - (Vmax * A / (Km + A)) / Volume, whereAis the amount. - Define initial estimates for THETA(1) (Vmax) and THETA(2) (Km) based on exploratory analysis (e.g., Eadie-Hofstee plot).
- Specify an appropriate residual error model (e.g., proportional, additive, or combined).
- Use the
SAEMorFOCEestimation method for stability and accuracy [20].
- Use
- Model Estimation: Execute the NONMEM control stream. Monitor the minimization and covariance steps for successful completion.
- Model Diagnostics: Evaluate goodness-of-fit: Visual Predictive Check (VPC), individual fits, residual plots. Assess precision of parameter estimates (relative standard error).
- Validation: Perform a bootstrap analysis (e.g., 1000 samples) using PsN to obtain nonparametric confidence intervals for Vmax and Km, verifying robustness.
Protocol 2: Implementing the Enhanced OptiDose Algorithm in NONMEM
This protocol details the translation of a validated PKPD model into an optimized dosing regimen using the enhanced OptiDose method [61].
Objective: To compute an individualized dosing regimen that minimizes a primary efficacy cost function while strictly adhering to a secondary safety constraint.
Materials:
- A finalized population PKPD model with fixed parameters (e.g., from Protocol 1 and subsequent PKPD modeling).
- Software: NONMEM with compiler support, OptiDose algorithm templates [61].
Procedure:
- Problem Formulation:
- Control Stream Configuration:
- Declare doses (
D1, D2,...) as parameters to be estimated (THETA). - Implement the penalty method by augmenting the cost function:
J_total = J + (ρ/2) * INTEGRAL( max(0, g(y))² )[61]. - Use the
L-BFGS-Bor other constrained optimization algorithm via$ESTIMATION MAXEVALS=9999 METHOD=CONDITIONAL.
- Declare doses (
- Execution & Iteration:
- Run NONMEM to solve the unconstrained optimization problem.
- Check constraint violation. If the constraint is violated, increase the penalty parameter (ρ) and re-run until the solution is feasible.
- Verify optimality by performing a local sensitivity analysis around the optimal doses.
Protocol 3: High-Performance Computing (HPC) Execution for Large-Scale Optimization
For complex models or population-level optimization, HPC resources are essential [62].
Objective: To efficiently execute computationally intensive NONMEM OptiDose runs using parallel processing on an HPC cluster (e.g., Metworx).
Materials: HPC cluster with SGE/Slurm, NONMEM installed with MPI support, model files.
Procedure:
- Environment Configuration: Configure your session to use compute nodes (e.g., 16-32 vCPU) rather than the head node [62].
- Parallel Job Submission:
- Using
bbr: Submit model with arguments.bbi_args = list(threads=16, parallel=TRUE)[62]. - Using PsN: Use the
-parafileoption with a.pnmfile specifyingNODES=16.
- Using
- Performance Tuning: Use empirical testing (e.g.,
bbr::test_threads()) to identify the optimal number of cores, balancing speed and resource utilization [62].
Visualizing Workflows and Logical Relationships
Diagram 1: PKPD Model Development & OptiDose Optimization Workflow (100 chars)
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Essential Toolkit for M-M Estimation and OptiDose Implementation
| Tool / Reagent Category | Specific Item / Software | Function in Research | Key Feature / Note |
|---|---|---|---|
| Primary Modeling Software | NONMEM 7.6 [20] | Gold-standard for NLME PKPD modeling, parameter estimation, and OptiDose algorithm execution. | Includes SAEM, BAYES, advanced solvers (ADVAN16), and parallel computing support. |
| HPC & Parallelization | Metworx Platform [62] | Cloud-based environment for executing large, parallelized NONMEM jobs efficiently. | Enables configuration of compute nodes matching core counts for optimal parallel speed. |
| Model Execution Interfaces | bbr R package [62], PsN [62], Pirana [62] |
Streamline model submission, management, and execution, especially on HPC clusters. | bbr::test_threads() helps empirically determine optimal core count for parallel runs. |
| Regulatory & Strategic Framework | FDA Project Optimus Guidance [60] | Provides regulatory framework and expectations for dose optimization in oncology. | Shifts paradigm from MTD to OBD, justifying the need for OptiDose-type approaches. |
| Model Diagnostics & Evaluation | Nonparametric Estimation (NONP) in NONMEM [26] | Evaluates and identifies non-normal parameter distributions without restrictive assumptions. | Corrects bias from FO method; crucial for accurate variability estimation for simulation. |
| Advanced Modeling Techniques | Fractional Differential Equation (FDE) Subroutine [17] | Extends NONMEM to model systems with memory/anomalous kinetics using FDEs. | Useful for complex PK where traditional compartment models fail. |
| Trial Design & Dose Selection | Optimal Biological Dose (OBD) Concept [59] | The target clinical dose that optimally balances efficacy and toxicity. | The clinical endpoint of the OptiDose optimization process. |
Solving Convergence Issues and Optimizing Michaelis-Menten Model Performance
Within the context of Michaelis-Menten (MM) parameter estimation using NONMEM, model instability represents a critical barrier to obtaining reliable, reproducible, and biologically plausible estimates of Vmax and Km. Instability is not a singular error but a symptomatic outcome of deeper issues in the interaction between the model structure, the data, and the estimation algorithm [43]. For MM kinetics, which are inherently nonlinear, these issues are often exacerbated. A stable model reliably converges to the same solution with minimal sensitivity to initial estimates, yielding precise parameter uncertainties. An unstable model manifests through failed minimization runs, lack of convergence, biologically implausible parameter estimates (e.g., Vmax approaching infinity), or unreportable standard errors [43]. This document outlines a systematic, practical framework for diagnosing the root causes of instability—focusing on failed minimization and parameter uncertainty—and provides protocols to resolve them, ensuring robust MM parameter estimation.
Core Diagnostic Framework: Symptoms and Root Causes
The first step in diagnosing instability is to categorize the observed symptoms and link them to their most probable underlying causes. This facilitates a targeted investigative approach.
Table 1: Diagnostic Matrix of Model Instability Symptoms and Probable Causes in NONMEM [43]
| Symptom Category | Specific NONMEM Manifestations | Associated Probable Causes | Relevance to MM Kinetics |
|---|---|---|---|
| Minimization Failure | Run terminates with error (e.g., rounding error). Failure to converge within iteration limits. | Over-parameterized model relative to data. Poor initial parameter estimates. Extreme correlation between parameters (e.g., Vmax and Km). | High correlation between Vmax and Km is common, especially with limited data spread across the nonlinear curve. |
| Uncertainty & Reliability | Convergence without standard errors (COVARIANCE STEP OMITTED). Large condition number. Different estimates from different initial values. | Insufficient data information content. "Flat" objective function region near minimum. Unidentifiable parameters. | Data may lack informative points near the Km, making its estimation highly uncertain and the OFV "flat". |
| Numerical & Performance | Zero gradients during search. Run does not stop (hanging). Poor mixing in Bayesian chains (BAYES). | Poorly scaled parameters (e.g., Km=0.001 vs. Vmax=100). Censored data (BLQ) handling issues [63]. | MM parameters often differ by orders of magnitude; improper scaling hinders gradient searches. BLQ data is common in PK. |
| Biological Implausibility | Theta estimates outside physiological/pharmacological bounds (e.g., negative Km, clearance > cardiac output). | Model misspecification. Data quality issues (outliers, mis-assigned doses). Local minima trap. | May indicate the model is trying to fit noise or that an alternative model (e.g., with parallel linear clearance) is needed. |
Detailed Experimental Protocols for Diagnosis and Resolution
The following protocols provide a step-by-step methodology for implementing the diagnostic workflow, specifically tailored for MM model development in NONMEM.
Protocol 1: Systematic Diagnosis of Instability
Objective: To isolate the root cause of minimization failure or excessive parameter uncertainty in a MM model. Workflow: Follow the logic of the diagnostic diagram (Section 5.1).
- Initial Check & Confirmation: Verify the model code accurately represents the intended structural model (e.g.,
$DESblock correctly codes the MM equationdA/dt = - (Vmax * A/V) / (Km + A/V)). Confirm data integrity (doses, concentrations, BLQ markers) [43]. - Evaluate Data Quality & Information Content:
- Plot observed concentration-time data per individual. For MM elimination, assess if data spans the informative nonlinear region (concentrations around and below Km).
- Quantify the proportion of BLQ data. Implement and compare different BLQ handling methods (M1, M3, M6+) [63]. The M3 method is precise but can cause instability; the M7+ method (imputing zero with inflated residual error) offers a stable alternative during development [63].
- Profile the Objective Function:
- Fix the parameter in question (e.g., Km) to a range of values (e.g., from 0.1 to 5 times the estimated value).
- For each fixed value, estimate all other parameters and record the final Objective Function Value (OFV).
- Plot OFV vs. parameter value. A flat profile indicates poor data information and unidentifiability. A sharp, well-defined minimum indicates good information but potential for correlation if profiles for Vmax and Km are correlated.
- Assess Parameter Correlation & Scaling:
- From a successful run, examine the correlation matrix from the covariance step. Correlation between Vmax and Km > |0.95| indicates a problematic, nearly non-identifiable relationship.
- Check parameter scaling: Initial estimates for parameters should be within a few orders of magnitude of each other. Rescale parameters (e.g., estimate Km in mg/L instead of ng/mL) to improve optimizer performance.
Protocol 2: Generating Robust Initial Estimates for MM Parameters
Objective: To automate the generation of stable initial estimates to prevent minimization failure, using a data-driven pipeline [64]. Background: Poor initial estimates are a primary cause of failed convergence. This protocol adapts a generalized pipeline for use with MM kinetics.
- Data Preparation: Pool data naively (treating all data as from a single subject) based on time after dose (TAD). Bin and calculate median concentrations per time window [64].
- Visual-Guided Estimation (Graphic Method):
- For IV bolus data with MM elimination, plot log(Concentration) vs. time. The terminal linear phase slope estimates
-Vmax/(Km*V). The y-intercept helps estimate V. - Use the integrated form of the MM equation to perform nonlinear regression on the pooled data to get initial
VmaxandKmestimates.
- For IV bolus data with MM elimination, plot log(Concentration) vs. time. The terminal linear phase slope estimates
- Parameter Sweeping for Refinement:
- Define plausible ranges for
VmaxandKm(e.g.,Vmax± 3-fold,Km± 10-fold from the graphic method estimate). - Use a grid search or stochastic search to evaluate multiple (
Vmax,Km) pairs. For each pair, simulate the model and compute the relative Root Mean Squared Error (rRMSE) against the pooled median data [64]. - Select the parameter pair yielding the lowest rRMSE as the robust initial estimate for the population model.
- Define plausible ranges for
Protocol 3: Resolving Instability via Model Simplification & Re-parameterization
Objective: To stabilize an over-parameterized or poorly identifiable MM model.
- Simplify the Statistical Model:
- If IIV cannot be reliably estimated on both Vmax and Km, apply IIV to only the more informed parameter (typically Vmax) and fix the other's omega to zero.
- Simplify the residual error model (e.g., from combined additive+proportional to proportional alone).
- Apply a Stabilizing Re-parameterization:
- For high correlation between Vmax and Km, re-parameterize the model. Instead of estimating
VmaxandKm, estimateVmaxandCLlin, whereCLlin = Vmax / Kmrepresents the linear clearance at very low concentrations. The MM equation becomes:Rate = (Vmax * C) / ( (Vmax/CLlin) + C ). This can dramatically reduce correlation.
- For high correlation between Vmax and Km, re-parameterize the model. Instead of estimating
- Incorporate Prior Information (Bayesian Estimation):
Table 2: Research Reagent Solutions for Stable NONMEM Modeling [43] [63] [20]
| Item Name | Function & Purpose | Key Considerations for MM Models |
|---|---|---|
| Automated Initial Estimate Pipeline [64] | An R-based tool to generate data-driven initial estimates for structural and statistical parameters, reducing manual effort and guesswork. | Crucial for MM models to provide starting points that respect the nonlinear saturation curve, preventing early minimization failure. |
| BLQ Data Handler (M7+ Method) [63] | A pragmatic method for BLQ data: impute zero concentration and inflate the additive residual error for these points. Balances stability with acceptable bias. | Provides a stable alternative to the exact but often unstable M3 method during MM model development, especially with sparse PK data. |
| Parallel Computing Configuration [62] | Guidelines for configuring NONMEM to run on high-performance computing (HPC) grids (e.g., Metworx) using bbr, PsN, or Pirana with threads=N. |
Essential for running complex MM models with large datasets, robust profiling, or Bayesian (BAYES) analysis, which are computationally intensive. |
| Model Diagnostic Suite (bbr/PsN) | Open-source R package (bbr) and Perl toolkit (PsN) for managing model runs, conducting bootstrap, and performing VPC. |
The test_threads() function in bbr helps empirically determine the optimal cores for parallelization, improving efficiency [62]. |
| ADVAN13/ADVAN16 (NONMEM Subroutines) [20] | PREDPP subroutines for solving general nonlinear models (ADVAN13) and stiff delay differential equations (ADVAN16). |
ADVAN13 can be used to directly code MM kinetics via differential equations, offering flexibility for complex disposition models. |
Visual Guides to the Diagnostic and Resolution Workflow
Diagnostic Decision Pathway for Model Instability
Diagram 1: A logical workflow for diagnosing the root cause of model instability, guiding the user from symptoms to actionable resolution strategies.
Pipeline for Generating Initial Parameter Estimates
Diagram 2: An automated, data-driven pipeline for generating robust initial parameter estimates to ensure stable model start-up [64].
Application Notes
Within the context of advanced pharmacokinetic (PK) research using NONMEM, the Michaelis-Menten (M-M) model is a cornerstone for describing saturable elimination processes (e.g., metabolism via CYP enzymes). A critical challenge is selecting a model that is sufficiently complex to capture the underlying biology without being overparameterized for the available data, which leads to poor parameter identifiability and predictive performance.
Table 1: Model Selection Criteria for Michaelis-Menten Analysis in NONMEM
| Criterion | Formula (Approx. in NONMEM) | Interpretation in Context | Optimal Choice |
|---|---|---|---|
| Objective Function Value (OFV) | -2 * Log(Likelihood) | Used for nested model comparison. A drop of >3.84 (χ², p<0.05, 1 df) suggests significant improvement. | Lower is better for nested models. |
| Akaike Information Criterion (AIC) | OFV + 2 * P | Penalizes for number of parameters (P). Balances fit and complexity for non-nested models. | Lower value indicates better balance. |
| Bayesian Information Criterion (BIC) | OFV + P * log(N) | Stronger penalty than AIC, dependent on sample size (N). Favors simpler models. | Lower value indicates better balance. |
| Relative Standard Error (RSE%) | (SE/Estimate)*100 | Precision of parameter estimates (Vmax, Km). RSE >30-50% indicates poor identifiability. | Lower RSE indicates higher confidence. |
| Visual Predictive Check (VPC) | Graphical comparison of observed vs. model-simulated percentiles. | Assesses model's predictive performance across the data range. | Observed percentiles within simulated confidence intervals. |
Table 2: Typical Scenarios & Root Causes of Poor Identifiability in M-M Estimation
| Scenario | Symptom (in NONMEM Output) | Probable Root Cause | Remedial Action |
|---|---|---|---|
| Sparse data near Km | High RSE for Km, correlation >0.9 between Vmax and Km. | Data information content insufficient to identify the inflection point. | 1. Collect richer sampling around expected Km. 2. Consider a simpler linear model if only ascending limb is observed. |
| High dose saturation | High RSE for Vmax, successful estimation of Km. | Lack of data in the linear, non-saturated range to characterize the maximum rate. | Incorporate lower dose data to define the initial slope. |
| Complex model on sparse data | Failure to converge, or covariance step omitted. | Model complexity (e.g., + covariates, inter-occasion variability) exceeds data information. | Simplify model hierarchically: remove covariates, then random effects. |
| Misspecified residual error | Systemic biases in Conditional Weighted Residuals (CWRES) vs. time/prediction. | Error model (e.g., constant CV vs. additive) mis-specified, distorting structural parameter estimates. | Test alternative error models, use extended least squares. |
Experimental Protocols
Protocol 1: Base Michaelis-Menten Model Development in NONMEM Objective: To develop a population PK model for a drug exhibiting saturable elimination using NONMEM.
- Data Preparation: Structure dataset with columns: ID, TIME, AMT, DV (observed concentration), EVID (event identifier), RATE for infusion, and relevant covariates (e.g., WT, CYP2D6 phenotype). Ensure doses (AMT) span a range expected to produce both linear and saturated elimination.
- Model Specification:
- Structural Model: Use the ADVAN13 $DES block or the analytical solution (ADVAN2) to code the M-M equation: dA/dt = - (Vmax * A / (Km + A)) / V, where A is amount in the central compartment, V is volume of distribution.
- Inter-Individual Variability (IIV): Apply exponential error models to Vmax and Km (e.g.,
Vmax = TVVMAX * EXP(ETA(1))). Estimate OMEGA matrix. - Residual Error Model: Start with a proportional (constant CV) error model:
DV = IPRED * (1 + EPS(1)).
- Estimation: Use the First-Order Conditional Estimation (FOCE) with INTERACTION or the more robust Stochastic Approximation Expectation-Maximization (SAEM) method for initial estimates.
- Diagnostics: Generate standard goodness-of-fit plots (Observed vs. Individual/Population Predictions, CWRES vs. TIME/PRED) and evaluate parameter precision (RSE%).
Protocol 2: Stepwise Covariate Model Building on M-M Parameters Objective: To identify significant demographic/pathophysiological covariates on Vmax and Km.
- Base Model: Establish a converged model with IIV on Vmax and Km from Protocol 1.
- Forward Inclusion: a. Graph individual ETA estimates for Vmax and Km vs. potential covariates (e.g., body weight, renal function). b. For each plausible covariate-parameter relationship (linear, power, exponential), add it to the model one at a time. c. Use the Likelihood Ratio Test (LRT): a drop in OFV > 6.64 (χ², p<0.01, 1 df) signifies significance for forward inclusion.
- Backward Elimination: a. After all significant covariates are included, refit the full model. b. Remove each covariate one by one and increase the OFV. Use a stricter criterion (e.g., OFV increase > 10.83, p<0.001) to retain the covariate in the final model.
- Final Evaluation: Perform a Visual Predictive Check (VPC) stratified by key covariates (e.g., renal impairment status) to validate the final model's predictive performance across subpopulations.
Mandatory Visualizations
Title: NONMEM M-M Covariate Model Building Workflow
Title: Data Information vs. Model Complexity Balance
The Scientist's Toolkit: Essential Reagents & Software for M-M NONMEM Analysis
| Item | Category | Function/Brief Explanation |
|---|---|---|
| NONMEM | Software | Industry-standard software for nonlinear mixed-effects modeling of PK/PD data. |
| PsN (Perl-speaks-NONMEM) | Software Toolkit | Provides essential utilities for automated model management, bootstrapping, VPC, and cross-validation. |
| Xpose or Pirana | GUI & Diagnostics | Facilitates model diagnostics, result management, and generation of diagnostic plots. |
| R or Python | Programming Language | Used for data preparation, post-processing of NONMEM outputs, and custom graphics. |
| PDx-Pop or Monolix | Alternative Software | Used for model validation or comparison using different estimation algorithms (e.g., SAEM in Monolix). |
| Rich PK Sampling Strategy | Experimental Design | Data collected across a dose range spanning linear and saturated elimination is crucial for M-M identifiability. |
| CYP Phenotype/Genuotype Data | Covariate | Key categorical covariate for Vmax in models of drugs metabolized by polymorphic enzymes. |
| Body Size Metrics (e.g., BSA) | Covariate | Continuous covariate commonly tested on Vmax (allometric scaling) and volume of distribution. |
Abstract Within the framework of NONMEM-based Michaelis-Menten parameter estimation research, achieving reliable and computationally stable parameter estimates is a fundamental challenge. This article presents a set of practical methodologies—reparameterization, parameter scaling, and the application of boundary constraints—to address common issues in nonlinear mixed-effects modeling. We provide detailed application notes and experimental protocols, demonstrating how these techniques enhance model identifiability, improve estimation algorithm performance, and ensure the biological plausibility of parameter estimates for maximum reaction rate (Vmax) and Michaelis constant (Km). The protocols are contextualized within the NONMEM 7.6 environment [20] and illustrated using the PREDPP ADVAN10 subroutine, which is explicitly designed for Michaelis-Menten elimination kinetics [19] [36].
The Michaelis-Menten equation is pivotal for modeling saturable metabolic processes in pharmacokinetics. However, estimating its parameters, Vmax and Km, from typical sparse, noisy clinical data presents significant difficulties. The parameters are often highly correlated, and the estimation surface can be flat, leading to poor identifiability, convergence failures, or biologically implausible estimates (e.g., negative values) [24]. These problems are exacerbated in population models where inter-individual variability must be quantified.
This article, framed within a broader thesis on advanced estimation techniques in NONMEM, addresses these challenges through three foundational "practical fixes." The focus is not on introducing novel statistical theory but on providing actionable, detailed protocols for implementing established numerical stabilization techniques within the NONMEM framework. Proper application of these methods is critical for robust population pharmacokinetic-pharmacodynamic (PK/PD) analysis, which forms the basis for model-informed drug development decisions [20] [54].
Reparameterization for Improved Identifiability and Stability
Reparameterization involves rewriting the model's parameterization to create a form that is more amenable to estimation. For the Michaelis-Menten model, this often aims to reduce the correlation between parameters and to define parameters with more orthogonal influences on the objective function.
1.1 Theoretical Foundation
The standard Michaelis-Menten equation for elimination rate (V) is:
V = (Vmax * C) / (Km + C), where C is the substrate concentration.
A common and effective reparameterization is to express the model in terms of intrinsic clearance (CLint) and Vmax, where CLint = Vmax / Km. An alternative parameter, such as Km, is retained. The equation becomes:
V = (Vmax * C) / (Km + C) = (CLint * Km * C) / (Km + C).
This transformation can help when Vmax and Km are highly correlated, as CLint and Km may have a weaker correlation. Another advanced reparameterization uses a reference concentration (Cref) and the fractional saturation at that concentration to define parameters that are more directly interpretable and better scaled.
1.2 NONMEM Implementation Protocol The following protocol details the implementation of a CLint-reparameterized Michaelis-Menten model using the PREDPP ADVAN10 subroutine.
Protocol 1.1: Implementing a CLint-Reparameterized Model in NM-TRAN
- Objective: To specify a one-compartment model with Michaelis-Menten elimination in NONMEM using a CLint and Km parameterization to improve estimation stability.
- Software: NONMEM 7.6 [20], PREDPP Library (ADVAN10) [19] [36].
Control Stream Key Records:
Procedure:
- Prepare a dataset (
data.csv) with required columns:ID,TIME,AMT,DV(drug concentration),CMT,EVID, andMDV[19] [65]. - Implement the control stream as shown. The
$PKblock defines the typical values (TVCLINT,TVKM), calculatesTVVMAX, and applies exponential random effects (ETA) toCLINTandKM. - The individual
VMAXis derived as the product of individualCLINTandKM. - The model parameters
VMandKMMare assigned for ADVAN10 [19]. - Initial estimates for
$THETAshould be informed by prior knowledge or exploratory analysis [66]. - Execute the model using a command such as
nmfe76 run.ctl run.lstor through an interface like Pirana or Wings for NONMEM [19] [22].
- Prepare a dataset (
- Diagnostic & Interpretation: Successful reparameterization often yields a reduced correlation between the estimated ETAs for CLint and Km (visible in the
$COVARIANCEoutput) and more stable convergence. Compare the condition number of the covariance matrix or the correlation matrix of parameter estimates with the standard parameterization to assess improvement.
Table 1: Impact of Reparameterization on Parameter Correlation (Simulated Example)
| Parameterization | Estimated Parameter Pair | Correlation Coefficient | Comment |
|---|---|---|---|
| Standard | ETA(Vmax) vs. ETA(Km) | 0.95 | High correlation, potential identifiability issues. |
| CLint-based | ETA(CLint) vs. ETA(Km) | 0.65 | Moderately correlated, improved stability. |
Parameter Scaling for Optimized Algorithm Performance
Parameter scaling ensures all parameters subject to estimation have similar orders of magnitude. This is crucial because gradient-based estimation algorithms can fail or become inefficient when parameters differ by several log orders, as is common in PK models (e.g., a Km of 1.2 mg/L vs. a Vmax of 1200 mg/h).
2.1 Principle of Scaling The goal is to transform parameters so their initial estimates and typical values are close to 1 (or within an order of magnitude of 1). This is typically done by dividing the parameter by a scaling constant (e.g., a population typical value or a convenient unit).
2.2 NONMEM Scaling Protocol
This protocol integrates scaling directly into the $PK block.
Protocol 2.1: Implementing Parameter Scaling in the Control Stream
- Objective: To scale the parameters Km and Vmax to improve the numerical behavior of the estimation algorithm (e.g., FOCE, SAEM).
- Method: Internal scaling within the control stream.
Control Stream Modifications (
$PKBlock):Procedure:
- Choose scaling constants (
SCALE_xxx) that reflect reasonable prior estimates of the parameters. - Define the scaled typical values (
TVxxx_SCALED) asTHETAs. With initialTHETAestimates fixed at 1, the initial biological parameter values equal the scaling constants. - In the
$THETArecord, initial estimates are set to 1. TheFIXoption can be used during initial testing to ensure the scaling works as intended before allowing estimation. - After ensuring the model runs, remove the
FIXed status and provide bounded initial estimates around 1 (e.g.,(0.5, 1, 5)). - The final estimated
THETAvalues are multiplied by the scaling constants to interpret the results in biological units.
- Choose scaling constants (
- Diagnostic & Interpretation: Improved algorithm performance is indicated by faster convergence, fewer minimization failures, and more accurate calculation of the covariance matrix. The scaled thetas (close to 1) lead to a better-conditioned Hessian matrix.
Table 2: Example of Parameter Scaling for a Michaelis-Menten Model
| Parameter (Biological) | Typical Value | Scaling Constant | Scaled Theta (Initial) | Scaled Theta Bounds |
|---|---|---|---|---|
| Michaelis Constant (Km) | 2.5 mg/L | 2.5 | THETA(1) = 1.0 | (0.1, 1.0, 10.0) |
| Max. Velocity (Vmax) | 50 mg/h | 50 | THETA(2) = 1.0 | (0.2, 1.0, 20.0) |
Boundary Constraints to Ensure Biological Plausibility
Boundary constraints prevent parameters from taking on nonsensical values (e.g., negative clearances or volumes) during estimation, which can cause numerical overflow or crash. They incorporate prior knowledge into the estimation.
3.1 Types of Constraints in NONMEM
NONMEM allows bounds to be set on $THETA (population parameters) and $OMEGA/$SIGMA$ (variance components). For variance components, a lower bound of a small positive number is common to avoid estimations approaching zero, which can complicate covariance step calculations [67].
3.2 Protocol for Applying Boundaries
This protocol combines bounding THETA with the use of the NOABORT option and $ESTIMATION settings for robust estimation.
Protocol 3.1: Implementing Bounded Estimation for Michaelis-Menten Parameters
- Objective: To constrain population parameter (THETA) and variance (OMEGA) estimates to biologically plausible ranges and ensure a successful covariance step.
Control Stream Modifications:
Procedure:
- Set lower and upper bounds on
$THETAusing the format(lower, initial, upper). The lower bound for kinetic parameters should always be >0. - For variance components (
$OMEGA,$SIGMA), a lower bound can be implicitly provided using a positive initial estimate. To set an explicit lower bound, use the format(0.01)for an initial estimate of 0.01 with a lower bound of 0 [67]. TheSAMEoption can be used for variance blocks. - Use the
NOABORToption in$ESTIMATIONto prevent the run from stopping if a numerical error occurs during the covariance calculation, allowing for troubleshooting. - The
MAXEVALoption prevents indefinite looping. - If boundary estimates occur (parameters "pin" at a boundary), this indicates model misspecification, lack of information in the data, or that the bound may be too restrictive.
- Set lower and upper bounds on
- Diagnostic & Interpretation: Check the final parameter estimates in the output file. Parameters hitting a bound will be flagged. Investigate such outcomes by reviewing the data, model structure, or relaxing the bounds if justified. A successful covariance step (
$COVARIANCE) with printed standard errors indicates that the estimation was numerically stable near the final estimates.
Integrated Experimental Protocol for Michaelis-Menten Model Development
This protocol integrates the three practical fixes into a coherent workflow for developing a population Michaelis-Menten model, following established model-building stages [66].
- Protocol 4.1: End-to-End Model Development with Practical Fixes
- Phase 1: Data Preparation and Exploratory Analysis
- Phase 2: Base Model Development
- Start Simple: Implement a base model using ADVAN10/TRANS1 with a CLint-Km reparameterization (Protocol 1.1).
- Apply Scaling: Incorporate parameter scaling (Protocol 2.1) using the initial estimates from Phase 1.
- Apply Constraints: Set liberal but biologically plausible bounds on
$THETAand$OMEGA(Protocol 3.1). - Initial Estimation: Run with a simple method (e.g.,
METHOD=CONDITIONALINTER [67]) to obtain preliminary estimates. - Diagnose: Evaluate goodness-of-fit plots (e.g., DV vs. PRED, CWRES vs. TIME) [66].
- Phase 3: Model Refinement & Final Estimation
- Refine the statistical model (covariate relationships, variance models) based on diagnostics.
- Perform final parameter estimation using a more rigorous method (e.g.,
METHOD=SAEMfollowed byMETHOD=IMP) [20]. - Execute the final
$COVARIANCEstep and verify that no parameters are at boundaries and that standard errors are reasonable. - Use the
$DESIGNfeature prospectively to evaluate the expected precision of parameter estimates (e.g., Vmax, Km) for the design used, or retrospectively to understand estimability [54].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Software and Components for NONMEM-based Michaelis-Menten Research
| Item | Function / Purpose | Example / Note |
|---|---|---|
| NONMEM 7.6 | Core engine for nonlinear mixed-effects model estimation [20]. | Industry standard software; includes PREDPP and NM-TRAN [20]. |
| PREDPP Library - ADVAN10 | Pre-built subroutine for one-compartment models with Michaelis-Menten elimination [19] [36]. | Used with $SUBROUTINES. TRANS1 provides the standard VM, KM parameterization [19]. |
| NM-TRAN Control Stream | User-written instruction file specifying the model, parameters, data, estimation methods, and output [65]. | A plain text file (.ctl) containing $PROBLEM, $INPUT, $PK, $THETA, etc. [65]. |
| Fortran Compiler | Compiles the NONMEM and user-supplied Fortran code into an executable [20]. | Required for installation. Intel Fortran Compiler or gFortran are common [20]. |
| Interface / Wrapper Software | Facilitates run management, model tracking, and graphical diagnostics [19]. | Pirana, Perl Speaks NONMEM (PsN), Wings for NONMEM, or R-based tools like xpose [19]. |
| Data File (.csv) | Contains all individual subject data in a structured format for NM-TRAN to read [19] [22]. | Required columns include ID, TIME, AMT, DV, EVID, CMT, MDV [19]. |
| Optimal Design Evaluator | Evaluates or optimizes study design using the Fisher Information Matrix (FIM) [54]. | NONMEM's $DESIGN record or external tools like PopED and PFIM [54]. |
Visual Appendix: Workflow and Model Diagrams
Diagram 1: Integrated model-building workflow incorporating the three practical fixes.
Diagram 2: Information flow within the NONMEM system [65].
Diagram 3: Schematic of Michaelis-Menten saturable elimination from a central compartment, as modeled by ADVAN10.
Leveraging Automated Pipelines and Tools for Initial Estimate Generation and Model Search
This document provides detailed application notes and protocols for implementing automated workflows within NONMEM-based research, specifically contextualized within a broader thesis focusing on Michaelis-Menten parameter estimation. Nonlinear mixed-effects (NLME) modeling, the foundation of population pharmacokinetic (PopPK) analysis, is critically dependent on two resource-intensive processes: obtaining robust initial parameter estimates and conducting a thorough search for the optimal model structure. Manual approaches to these tasks are time-consuming, require expert judgment, and can lead to suboptimal models due to local minima or investigator bias [68] [69].
Recent advancements have produced automated pipelines for initial estimate generation that are effective even with sparse data [7] and intelligent global search algorithms that systematically explore vast model spaces [68] [69]. Integrating these tools creates a powerful, standardized framework for pharmacometric analysis. For research centered on Michaelis-Menten kinetics—a system inherently nonlinear and often challenging to fit—leveraging such automation is particularly valuable. It ensures a reproducible, rigorous approach to model development, allowing the researcher to focus on the biological interpretation of the saturated enzymatic process rather than the intricacies of model tuning and search logistics.
Automated Pipeline for Initial Estimate Generation
The generation of high-quality initial estimates is a prerequisite for successful and efficient model convergence in NONMEM. An automated, data-driven pipeline has been developed to replace manual methods like non-compartmental analysis (NCA), which struggles with sparse data [7].
Pipeline Architecture and Workflow
The pipeline is designed to process data formatted to nlmixr2 standards and proceeds through three consecutive parts to compute initial estimates for structural and statistical parameters [7].
Diagram 1: Architecture of the automated initial estimate pipeline [7].
Core Methodologies and Protocols
Protocol 2.2.1: Data Preparation and Naïve Pooling
- Objective: To standardize raw data and create pooled concentration-time profiles for population-level analysis.
- Steps:
- Assign Dosing Information: Process observation records to assign dosing events, identify administration routes (IV bolus, infusion, extravascular), and calculate time after the last dose (TAD).
- Categorize Data: Separate data into three groups: first-dose, non-first-dose (multiple-dose), and mixed-dose data.
- Pool and Bin: For each group, bin concentration-time data based on TAD. Use a default of 10 time windows (adjusting if fewer unique time points exist) to ensure adequate PK curve characterization.
- Calculate Representative Values: Within each time window, compute the median time and median drug concentration. These paired medians form the pooled concentration-time profile used in subsequent steps [7].
Protocol 2.2.2: Adaptive Single-Point Method
- Objective: To estimate clearance (CL) and volume of distribution (Vd) from datasets with sparse sampling (e.g., one sample per individual).
- Base Phase Protocol:
- Estimate Half-life: Perform linear regression on the terminal phase of the naïve pooled data to estimate elimination rate constant (λz) and half-life (t₁/₂).
- Calculate Vd: For intravenous data, identify the first concentration (C₁) sampled within 0.2*t₁/₂ after a first dose. Approximate Vd as Dose / C₁.
- Calculate CL: Under steady-state conditions (achieved after ≥5 doses), identify maximum (Cˢˢmax) and minimum (Cˢˢmin) concentrations within a dosing interval (τ). Calculate average concentration (Cˢˢavg). Compute CL as Dose / (Cˢˢavg * τ).
- Summarize Population Estimates: Calculate the geometric mean (with a 5% trim) of individual parameter estimates to derive robust population initial values [7].
Protocol 2.2.3: Parameter Sweeping for Complex Models
- Objective: To find initial estimates for parameters in complex models (e.g., Michaelis-Menten elimination, multi-compartment distribution) where direct calculation is not feasible.
- Steps:
- Define Search Range: Establish a biologically plausible range for the parameter in question (e.g., Vmax, Km).
- Generate Candidates: Create a vector of candidate values spanning the defined range.
- Simulate and Compare: For each candidate value, simulate the model and predict concentrations. Hold other parameters constant at preliminary estimates.
- Evaluate Fit: Calculate the relative Root Mean Squared Error (rRMSE) between the observed data (or pooled profile) and the predictions for each candidate.
- Select Best Estimate: Choose the candidate parameter value that yields the lowest rRMSE as the initial estimate [7].
Performance and Validation Data
The pipeline was rigorously validated using simulated and real-world datasets.
Table 1: Validation Performance of the Automated Initial Estimate Pipeline [7]
| Dataset Type | Number of Datasets | Performance Outcome | Key Metric |
|---|---|---|---|
| Simulated | 21 | Final parameter estimates were closely aligned with pre-set true values. | Successful convergence with accurate final estimates. |
| Real-Life | 13 | Final parameter estimates were closely aligned with established literature references for the drugs. | Production of biologically plausible, literature-backed models. |
| Overall | 34 | The pipeline performed well in all test cases for both rich and sparse data scenarios. | 100% success rate in providing usable initial estimates. |
Automated Tools for Population Model Search
Once a base model is initialized, identifying the optimal structural and covariate model is the next challenge. Automated global search algorithms overcome the limitations of traditional stepwise "hill-climbing" methods, which are prone to local minima [68].
Search Algorithm Theory and Implementation
Local vs. Global Search: Traditional stepwise addition/backward elimination is a local search "greedy algorithm." It evaluates models close to the current best model and moves in the direction of immediate improvement. Its success depends entirely on starting in a region "convex" to the global optimum, an assumption often false in complex PK/PD model spaces [68].
Global search algorithms evaluate models that may be very different from each other, allowing them to escape local minima. Key algorithms include:
- Genetic Algorithms (GA): Mimic natural selection. A population of models (individuals) undergoes selection, crossover (mating), and mutation over generations to evolve towards better fit [68].
- Gene Expression Programming (GEP): An extension of GA that evolves complex mathematical expressions or model structures, useful for discovering non-polynomial covariate relationships [70].
- Bayesian Optimization with Surrogate Models: Uses a probabilistic model (e.g., random forest) to predict the performance of untested models, guiding the search efficiently towards promising regions of the vast model space [69].
Diagram 2: Workflow of a genetic algorithm for automated model search [68] [69].
Integrated Protocol: Automated Model Search with pyDarwin
Protocol 3.2.1: Setup and Pre-processing
- Objective: To configure an automated model search for a PopPK dataset, such as one featuring Michaelis-Menten elimination.
- Tools: pyDarwin (optimization library), NONMEM as the estimation engine [69].
- Steps:
- Data Formatting: Ensure the dataset is formatted for NONMEM, including necessary covariates.
- Define Model Search Space: Specify the universe of possible models. For extravascular drugs, this may include:
- Compartments: 1- or 2-compartment distribution.
- Absorption Models: First-order, zero-order, parallel, or transit compartments.
- Elimination Models: Linear, Michaelis-Menten, or combined.
- Covariate Relationships: Potential continuous or categorical covariates on parameters (e.g., weight on Vd, age on CL).
- Specify Penalty Function: Define an objective function that balances goodness-of-fit with model plausibility. A recommended composite penalty includes:
- Akaike Information Criterion (AIC): Penalizes model complexity to avoid overfitting.
- Parameter Plausibility Term: Penalizes abnormal parameter values (e.g., very high/low inter-individual variability, high shrinkage, unrealistic volume estimates) [69].
Protocol 3.2.2: Execution of the Global Search
- Objective: To efficiently explore the pre-defined model space and identify the optimal structure.
- Steps:
- Initialization: pyDarwin generates an initial population of random model structures from the search space.
- Iterative Evaluation Loop: a. Fitness Evaluation: Each model in the population is run in NONMEM (or a surrogate evaluates its expected performance). The composite penalty function score is calculated. b. Selection & Evolution: Algorithms like GA or Bayesian Optimization select the fittest models, apply genetic operators (crossover, mutation) or probabilistic inference to propose a new, promising generation of models.
- Convergence: The search continues for a set number of generations or until the improvement in the best fitness score falls below a threshold.
- Output: The model with the optimal (lowest) penalty function score is selected as the final, recommended model structure [69].
Performance Data and Comparison
Table 2: Performance of Automated Model Search (pyDarwin) vs. Manual Development [69]
| Metric | Automated Search (pyDarwin) | Traditional Manual Search |
|---|---|---|
| Average Development Time | Less than 48 hours (in a 40-CPU, 40 GB RAM environment). | Often several days to weeks, depending on model complexity and analyst workload. |
| Search Efficiency | Evaluated fewer than 2.6% of the total models in a >12,000-model search space. | Typically explores a minuscule fraction of the possible model space due to time constraints. |
| Risk of Local Minima | Low. Global search algorithms (GA, Bayesian Optimization) are designed to escape local minima. | High. Stepwise "hill-climbing" is intrinsically susceptible to local minima [68]. |
| Reproducibility | High. The search is defined by an explicit, code-based penalty function and search space. | Variable. Depends on analyst expertise and subjective judgment, leading to potential irreproducibility [69]. |
| Model Quality Outcome | Identified model structures were comparable or very close to manually developed expert models for clinical datasets [69]. | Dependent on the skill and experience of the modeler; can be optimal but consistency is not guaranteed. |
The Scientist's Toolkit: Essential Research Reagents & Software
Table 3: Key Software Tools and Methodological Components for Automated NONMEM Workflows
| Tool/Component | Primary Function | Role in Automated Workflow | Source/Reference |
|---|---|---|---|
| NONMEM 7.6 | Industry-standard software for NLME population PK/PD analysis. Provides multiple estimation methods (FOCE, SAEM, IMP, BAYES). | Core estimation engine. All automated pipelines ultimately execute candidate models here for parameter estimation and objective function calculation. | [20] [32] |
| R Package for Initial Estimates | Open-source R package implementing the automated pipeline (data prep, single-point, NCA, graphic methods, sweeping). | Provides the initial estimates for THETAs (structural parameters) required to start model estimation, crucial for complex models like Michaelis-Menten. | [7] [71] |
| pyDarwin | A Python library for global optimization. Implements genetic algorithms, Bayesian optimization, and other search strategies. | Orchestrates the automated model search. It generates candidate model structures, manages their evaluation (often via NONMEM), and evolves the population. | [69] |
| nlmixr2 | An R package for NLME modeling that provides a unified interface and facilitates model specification and simulation. | Serves as a data standard and potential alternative/supplementary estimation environment. The initial estimate pipeline uses its data format [7]. | [7] |
| Composite Penalty Function | A user-defined function combining AIC (for complexity) and parameter plausibility checks (for biological realism). | Guides the automated model search by objectively scoring and ranking candidate models, replacing subjective analyst judgment during model selection [69]. | [69] |
| Gene Expression Programming (GEP) | An evolutionary algorithm specialized for discovering symbolic mathematical relationships. | Useful for automated covariate model building, particularly for identifying non-linear or non-polynomial covariate-parameter relationships [70]. | [70] |
| Perl-speaks-NONMEM (PsN) | A versatile toolkit of Perl scripts for facilitating NONMEM modeling, including automated stepwise covariate analysis (SCM). | Represents an earlier generation of automation tools, primarily for covariate selection. Useful for specific, well-defined sub-tasks [72]. | [72] |
Within the domain of pharmacometrics and model-informed drug development, constructing robust nonlinear mixed-effects (NLME) models is fundamental. For research focused on Michaelis-Menten (M-M) parameter estimation using NONMEM, stability—defined as a model's reliable convergence to consistent and biologically plausible parameter estimates—is a critical yet frequently elusive goal [43]. Instability manifests as failed minimization, absence of standard errors, or parameter estimates that shift dramatically with minor changes in initial values or estimation algorithms [43] [32]. This undermines the reliability of key parameters like Vmax (maximum elimination rate) and Km (Michaelis constant), which are crucial for predicting nonlinear pharmacokinetics and dose-concentration relationships.
The root causes of instability are often multifaceted, arising from the intricate interplay between model complexity, data information content, and data quality [43]. An inherently complex M-M model may exceed what the available data can inform, leading to poorly identifiable parameters. Concurrently, common practices like suboptimal initial parameter estimates can trap estimation algorithms in local minima or cause premature termination [7]. This article presents a structured, heuristic workflow to diagnose and resolve these stability issues, framed explicitly within the context of advancing NONMEM-based M-M pharmacometric research.
Diagnostic and Resolution Workflow for Model Stability
The following workflow provides a systematic, step-by-step approach for diagnosing the root cause of instability and implementing targeted corrective actions. It synthesizes a problem-solving heuristic [43] with foundational principles of NONMEM estimation [32] [19] and modern automated techniques for initial estimation [7].
Diagram 1: Heuristic Workflow for Diagnosing and Resolving Model Instability (94 characters)
Detailed Experimental Protocols & Application Notes
Protocol 1: Generating Robust Initial Parameter Estimates for M-M Models
Poor initial estimates are a primary cause of convergence failure [7]. This protocol details an automated, data-driven pipeline to generate reliable starting values for M-M parameters (Vmax, Km), volumes (V), and residual error, applicable to both rich and sparse data scenarios.
- Data Preparation: Format the dataset with required columns:
ID,TIME,AMT,DV(observed concentration),EVID(event identifier), andCMT(compartment number). Prepare a naïve pooled dataset by binning concentration-time data from all subjects based on time-after-dose windows and calculating median values for each window [7]. - Estimation for One-Compartment Linear Proxy:
- Use the pooled data to estimate a linear one-compartment model with first-order elimination (CL, V) as an initial proxy. Employ an adaptive single-point method: Calculate apparent clearance (CL) at steady state using the average of maximum and minimum concentrations (
Css,avg) and the dosing interval (τ):CL = Dose / (Css,avg * τ). Estimate volume (V) from the first post-dose concentration, provided it is sampled early (within ~20% of the estimated half-life) [7]. - Alternatively, apply graphic methods (e.g., terminal slope regression for intravenous data) or naïve pooled Non-Compartmental Analysis (NCA) to the median pooled profile to obtain CL and V estimates [7].
- Use the pooled data to estimate a linear one-compartment model with first-order elimination (CL, V) as an initial proxy. Employ an adaptive single-point method: Calculate apparent clearance (CL) at steady state using the average of maximum and minimum concentrations (
- Initializing M-M Parameters (Vmax & Km):
- Use the CL estimate from Step 2 as an approximation of the linear clearance at low concentrations. Set the initial Km to a value roughly in the middle of the observed concentration range.
- Employ parameter sweeping: Define a plausible range for Vmax (e.g., 0.5 to 2 times the dose/hour) and Km (e.g., 0.1 to 10 times the median concentration). Simulate the M-M model across a grid of these values using the pooled data structure. Select the (Vmax, Km) pair that minimizes the relative Root Mean Square Error (rRMSE) between simulated and median observed concentrations [7].
- Initializing Statistical Parameters:
- For inter-individual variability (IIV, OMEGA), set pragmatic diagonal elements (e.g., 0.09 for 30% coefficient of variation) for CL, V, Vmax, and Km [7].
- For residual unexplained variability (RUV, SIGMA), use a data-driven estimate from the variance of the pooled profile or a fixed default (e.g., additive error of 10% of the median observation) [7].
Protocol 2: Diagnostic Evaluation of Model Stability
After obtaining initial estimates, run a simplified "restricted" model (e.g., M-M elimination only, no covariates, diagonal OMEGA) with a robust estimation method like FOCE or IMP [32]. Analyze these key diagnostics to pinpoint instability [43]:
- Condition Number & R Matrix: In NONMEM output, examine the eigenvalues of the correlation matrix (R matrix). Calculate the condition number (ratio of largest to smallest eigenvalue). A condition number > 1000 indicates severe ill-conditioning and parameter non-identifiability [43].
- Standard Error Reliability: Check for successful calculation of standard errors (SE) for all parameters. Parameters with extremely large relative SE (>50%) or a failure to compute SEs indicate poor practical identifiability.
- Objective Function Value (OFV) Stability: Run the model multiple times from different starting points (perturbed by ±10-20%). A stable model will converge to a nearly identical OFV; unstable models will yield different OFVs or fail.
Protocol 3: Parameterization & Estimation Strategies for M-M Models
Based on the diagnostics, apply targeted corrections:
- For High Condition Number/Ill-Conditioning: Reparameterize the model. For a one-compartment M-M model, use the standard clearance (CL) and volume (V) parameterization (
ADVAN10 TRANS2in NONMEM) instead of micro-constants (ADVAN10 TRANS1) [19]. If Km is poorly identifiable, consider fixing it to a literature value from in vitro studies or simplifying the model to linear elimination if supported by the data [43]. - For Unreliable Standard Errors or Run Failures: Switch the estimation algorithm. Move from a linearization method (FOCE) to a sampling-based method like Importance Sampling (IMP) or Stochastic Approximation EM (SAEM), which are more robust for nonlinear models and can provide better uncertainty estimates [32]. For complex models defined by ordinary differential equations (ODEs), ensure the appropriate ODE solver (
ADVAN13,ADVAN14) is specified [19]. - Iterative Model Simplification: If instability persists, systematically reduce model complexity. This may involve fixing IIV on less variable parameters (e.g., Km), switching from a full to a diagonal OMEGA block, or using a simpler residual error model.
Data Presentation: Comparative Analysis of Methods
Table 1: Performance of Estimation Methods for Nonlinear Models (e.g., M-M) [32] [13]
| Estimation Method | Key Principle | Computational Speed | Robustness for M-M | Best Use Case in Stability Workflow |
|---|---|---|---|---|
| FO (First Order) | Linearizes inter & intra-individual variability. | Very Fast | Low (biased for nonlinearity) | Not recommended for M-M models. |
| FOCE (First Order Conditional Estimation) | More accurate approximation than FO. | Fast | Moderate | Initial diagnostic runs with good initial estimates. |
| IMP (Importance Sampling) | Monte Carlo integration in "important" parameter region. | Slow | High | Final estimation after stabilization; obtains reliable standard errors. |
| SAEM (Stochastic Approximation EM) | Markov Chain Monte Carlo (MCMC) sampling. | Slow (but efficient for ODEs) | High | Complex models where FOCE fails; often requires final IMP step for covariance. |
| Nonlinear Regression (Pooled Data) | Fits naïve pooled data ignoring IIV. | Very Fast | Medium (for initial estimates) | Protocol 1: Generating initial Vmax and Km estimates. |
Table 2: Common Stability Diagnostics and Interpretations [43]
| Diagnostic Metric | Acceptable Range | Indication of Instability | Suggested Corrective Action |
|---|---|---|---|
| Condition Number | < 1000 | > 1000 indicates ill-conditioning. | Reparameterize model; fix correlated parameters. |
| Relative Standard Error (RSE) | < 30-50% for key parameters | RSE > 50% or "NaN". | Simplify model; switch to IMP/SAEM method [32]. |
| OFV Variation from Different Starts | ∆OFV < 0.1 | ∆OFV > 2.0. | Improve initial estimates (use Protocol 1). |
| Minimization Status | "MINIMIZATION SUCCESSFUL" | "MINIMIZATION TERMINATED". | Check data quality/outliers; review model code for errors. |
Table 3: Recommended Parameterization for M-M Models in NONMEM [19]
| Model Feature | Problematic Parameterization | Recommended Stable Parameterization | Rationale |
|---|---|---|---|
| Structural Model | Micro-rate constants (ADVAN10 TRANS1: K, V). |
Clearance/Volume (ADVAN10 TRANS2: CL, V). |
Reduces parameter correlation; more intuitive. |
| M-M Parameters | IIV on both Vmax and Km. | IIV only on Vmax (or CL); fix Km or assign low IIV. | Km is often poorly identifiable; reduces complexity. |
| Statistical Model | Full OMEGA block for CL & V. | Diagonal OMEGA matrix. | Avoids estimating covariance, improving stability. |
| Residual Error | Complex combined error early on. | Simple additive or proportional error. | Fewer parameters to estimate during stabilization. |
The Scientist's Toolkit: Essential Research Reagents & Solutions
Table 4: Key Software Tools and Methodological "Reagents" for Stability Resolution
| Tool / Method | Category | Primary Function in Stability Workflow | Reference / Source |
|---|---|---|---|
| NONMEM (with PREDPP) | Core Software | Industry-standard engine for NLME model fitting and simulation. | [19] |
nlmixr2autoinit R package |
Automation Tool | Implements Protocol 1 for automated, data-driven generation of initial parameter estimates. | [7] [73] |
| IMP & SAEM Algorithms | Estimation Method | Robust, sampling-based estimation methods for unstable models or obtaining reliable covariance (Protocol 3). | [32] |
| Condition Number & R-matrix | Diagnostic Metric | Key numerical diagnostic for identifying parameter non-identifiability and ill-conditioning. | [43] |
| Naïve Pooled Data Analysis | Modeling Technique | Creates a median profile for graphic methods/NCA to inform initial estimates for complex models. | [7] |
| Model Reparameterization | Modeling Strategy | Technique to reduce parameter correlation (e.g., using CL instead of K) to improve conditioning. | [43] [19] |
| Perl-speaks-NONMEM (PsN)/Pirana | Run Management | Facilitates efficient model execution, diagnostic scripting, and workflow management. | [74] |
Validating Model Predictions and Comparing NONMEM with Emerging AI/ML Methodologies
Visual Predictive Checks (VPC) and numerical metrics are cornerstone techniques for validating nonlinear mixed-effects models (NLMEMs), particularly in the context of Michaelis-Menten (MM) pharmacokinetic models estimated using NONMEM. Within a thesis focused on MM parameter estimation, these diagnostics are essential for assessing whether a model can adequately describe observed data and for detecting structural or stochastic model misspecification [75] [76]. A VPC works by comparing key percentiles (e.g., 10th, 50th, 90th) of observed data against prediction intervals derived from hundreds or thousands of simulated datasets generated from the candidate model [75] [76]. This graphical check evaluates if the model can reproduce the central tendency and variability of the observations. For MM models, which are inherently nonlinear and can present identifiability challenges, VPCs are complemented by numerical metrics to provide a robust, multi-faceted assessment of model performance and reliability [16].
Visual Predictive Check Methodologies and Protocols
Core VPC Workflow and Protocol
The generation of a standard VPC follows a defined sequence of simulation, binning, and comparison.
VPC Generation and Assessment Workflow
Protocol 2.1.1: Standard VPC Generation for an MM Model
- Model Simulation: Using the final estimated MM model parameters, simulate 500-1000 replicate datasets in NONMEM using the
$SIMULATIONblock or a tool like PsN. Each replicate must match the original study design (doses, sample times, covariates) [77] [76]. - Data Binning: For both observed and simulated data, group observations into intervals (bins) of the independent variable (typically time). The number of bins (e.g., 8-10) is a trade-off; too few lose detail, too many create noisy percentiles [75]. Bins can be defined by equal time intervals, equal number of observations, or automated methods (e.g., Jenks) [75] [78].
- Percentile Calculation: Within each bin, calculate the desired percentiles (e.g., 10th, 50th, 90th) for the observed data. For the simulated data, calculate the same percentiles for each individual replicate.
- Prediction Interval (PI) Calculation: From the collection of simulated percentiles across all replicates, calculate the median and a prediction interval (e.g., the 5th to 95th percentile) for each percentile line (e.g., the 90% PI for the simulated 50th percentile) [77].
- Plotting and Assessment: Generate the VPC plot. The x-axis is typically the independent variable (time), and the y-axis is the dependent variable (e.g., concentration). The plot displays:
- Shaded areas representing the simulated prediction intervals for each percentile.
- Solid lines representing the median of the simulated percentiles.
- Symbols or dashed lines representing the observed percentiles. A model is considered adequate if the observed percentiles generally fall within the corresponding simulated prediction intervals [75] [76].
Advanced VPC Techniques
For complex MM models or heterogeneous study designs, standard VPCs can be misleading. Advanced correction techniques are required.
Prediction-Corrected VPC (pcVPC): This method corrects for variation in population predictions (PRED) across bins, which is crucial when doses or covariates vary widely. It normalizes observed (DV) and simulated data using the formula: pcY = Y * (median(PRED_bin) / PRED_ind) [77] [75]. This removes the confounding effect of the independent variable, allowing for a clearer diagnosis of model misfit.
Regression-Based VPC: This approach eliminates the subjective and sometimes problematic step of binning. It uses Additive Quantile Regression (AQR) to smoothly estimate percentiles for observed and simulated data over the independent variable, and LOESS regression to calculate the expected population prediction for pcVPC [79]. This method is particularly useful for sparse or irregularly sampled data.
Reference-Corrected VPC (rcVPC): A recent advancement, the rcVPC addresses the unintuitive y-axis scaling of pcVPC plots. It normalizes observations and simulations by the population prediction from a user-defined reference scenario (e.g., a standard dose), making trends directly interpretable in the original units and facilitating communication of exposure-response relationships [80].
Table 2.2.1: Comparison of VPC Techniques for MM Model Validation
| VPC Type | Key Principle | Primary Use Case | Advantages | Key Considerations |
|---|---|---|---|---|
| Standard VPC | Compares binned percentiles of observed vs. simulated data. | Homogeneous study designs with consistent dosing and sampling. | Intuitive, simple to implement and explain. | Misleading with variable dosing/covariates. Binning strategy affects results. |
| Prediction-Corrected VPC (pcVPC) | Normalizes data by typical population prediction per bin [77] [75]. | Heterogeneous designs (varying doses, strong covariates). | Removes bias from design variability, more accurate diagnosis. | Y-axis values are normalized and less intuitive [80]. |
| Regression-Based VPC | Uses quantile regression (AQR) instead of binning [79]. | Sparse, irregular data where binning is problematic. | Eliminates binning bias, provides smooth percentile estimates. | Computationally more intensive, requires selection of smoothing parameters. |
| Reference-Corrected VPC (rcVPC) | Normalizes data by predictions from a user-defined reference scenario [80]. | Communicating model performance, exploring exposure-response. | Y-axis in original units, highly intuitive, good for communication. | Requires definition of an appropriate reference scenario. |
Decision Logic for Selecting VPC Technique
Practical Implementation Protocol
Protocol 2.3.1: Implementing a pcVPC using NONMEM/PsN and R/xpose This protocol is central to a thesis on MM model validation [77] [78].
- Simulation with PsN: Execute the PsN
vpccommand with the-predcorrargument to ensure the population prediction (PRED) column is included in the simulation output. Specify the number of replicates (e.g.,-samples=500). This can also be done within NONMEM's control stream using$SIMULATION. Data Processing in R: Use the
xposepackage to read the simulation output.Generate pcVPC: Use the
vpc_data()andvpc()functions, specifyingvpc_type = 'continuous'. Thepredcorroption is automatically handled if the PsN run included-predcorr.Stratification: If the model includes categorical covariates (e.g., genotype affecting
Vmax), use thestratifyargument to create separate VPC panels for each subgroup, ensuring the model fits all subpopulations adequately [78] [76].
Numerical Validation Metrics for Michaelis-Menten Models
While VPCs provide a powerful visual diagnostic, numerical metrics are critical for objective assessment, especially for complex MM models where parameter identifiability can be an issue [16].
Parameter Estimation Diagnostics
Table 3.1.1: Key Numerical Metrics for MM Model Validation
| Metric Category | Specific Metric | Interpretation for MM Models | Target/Threshold |
|---|---|---|---|
| Precision of Estimates | Relative Standard Error (RSE%) | RSE% = (SE/Estimate)*100. High RSE% (>30-50%) for Vmax or Km suggests poor identifiability [16]. |
< 30% (ideal), < 50% (acceptable) |
| Model Convergence | NONMEM Termination Status | Successful covariance step (MINIMIZATION SUCCESSFUL, COVARIANCE STEP PASSED). |
Required. |
Significant Digits (SIGDIG) |
A measure of estimation precision. SIGDIG=3 is traditional but may be insufficient for nonlinear models; ≥4-6 is more robust [81]. | ≥ 4 | |
| Goodness-of-Fit (GOF) | Objective Function Value (OFV) | Used for hypothesis testing between nested models (ΔOFV). Lower is better. | ΔOFV > -3.84 (p<0.05) for 1 parameter difference. |
| Condition Number | Ratio of largest to smallest eigenvalue of the correlation matrix. Indicates collinearity (e.g., between Vmax and Km). |
< 1000 | |
| Simulation-Based | Normalized Prediction Distribution Errors (NPDE) | A statistical test comparing the entire distribution of observations to simulations. Can detect misfit not visible in VPC percentiles. | Mean ≈ 0, Variance ≈ 1, p-value of tests > 0.05 |
Protocol for Assessing Identifiability and Robustness
MM parameters (Vmax, Km) can be statistically non-identifiable, meaning different parameter pairs can fit the data equally well [16].
Protocol 3.2.1: Identifiability and Stability Check
- Profile Likelihood Analysis:
- Fix one MM parameter (e.g.,
Km) to a range of values around the estimate. - Re-estimate all other model parameters and record the OFV for each fixed value.
- Plot OFV vs. the fixed parameter value. A sharp, V-shaped trough indicates good identifiability. A flat or shallow trough suggests poor identifiability.
- Fix one MM parameter (e.g.,
- Bootstrap Analysis:
- Perform non-parametric bootstrap (e.g., 1000 resamples) to generate a distribution of parameter estimates.
- Calculate the median and 95% confidence intervals from the bootstrap distributions for
VmaxandKm. - Compare the original estimates to the bootstrap medians and CIs. Large shifts or very wide CIs indicate instability and potential identifiability problems.
- Global Optimization Verification: To rule out convergence to a local minimum—a known risk with derivative-based methods like FOCE in NONMEM—use a global optimization algorithm as a verification step [16]. Tools like Particle Swarm Optimization (PSO) or its variants (LPSO) can be applied. If PSO finds a significantly lower OFV than NONMEM's FOCE/SAEM, the original solution is likely a local minimum, and estimation should be restarted with different initial estimates [16].
The Scientist's Toolkit for MM Model Validation
Table 4.1: Essential Software and Research Reagent Solutions
| Tool/Reagent | Category | Primary Function in Validation | Application Note |
|---|---|---|---|
| NONMEM | Core Estimation Software | Estimates population parameters (THETA, OMEGA, SIGMA) for MM models using FOCE, SAEM, or IMP methods [32]. | The workhorse for NLMEM estimation. Outputs model files and simulations for diagnostics. |
| Perl Speaks NONMEM (PsN) | Toolkit / Wrapper | Automates complex tasks: VPC simulation, bootstrap, model comparison [77] [78]. | Essential for running standardized, reproducible validation workflows. |
| R Statistical Environment | Data Analysis & Visualization | Data manipulation, statistical calculation, and generation of diagnostic plots (GOF, VPC) [77] [79] [78]. | The flexible, open-source platform for custom analysis and graphics. |
| xpose R Package | Dedicated Diagnostics Package | Specialized for reading NONMEM output and creating pharmacometric diagnostics, including VPCs [78] [82]. | Streamlines the creation of publication-quality VPC and GOF plots. |
| Monolix Suite | Alternative Software | Performs NLMEM estimation and provides integrated, advanced VPC features for various data types [75]. | Useful for cross-verification of NONMEM results and its VPC interface is user-friendly. |
| Particle Swarm Optimization (PSO) Code | Global Optimization Algorithm | Verifies that NONMEM estimates represent a global, not local, minimum of the objective function [16]. | Critical final check for MM models prone to identifiability issues. Custom or published code (e.g., LPSO) is required. |
| High-Performance Computing (HPC) Cluster | Computational Resource | Provides the necessary power to run large simulation tasks (1000+ VPC replicates, bootstrap). | Enables thorough validation within practical timeframes for complex models. |
Within pharmacometrics, Model-Informed Drug Development (MIDD) leverages mathematical models to optimize drug development and dosing strategies [83]. For decades, nonlinear mixed-effects modeling (NLMEM) implemented in software like NONMEM has been the gold standard for population pharmacokinetic (PPK) analysis, including the estimation of Michaelis-Menten (MM) parameters that characterize saturable metabolic or transport processes [83] [32]. These traditional methods are built on mechanistic principles, offering interpretability and regulatory familiarity.
The emergence of artificial intelligence and machine learning (AI/ML) presents a paradigm shift, promising enhancements in predictive accuracy, computational efficiency, and the ability to handle complex, high-dimensional data [83] [84]. This application note provides a structured, evidence-based comparison between traditional NONMEM MM models and contemporary AI/ML predictors. Framed within broader thesis research on MM parameter estimation, it details experimental protocols, quantitative benchmarks, and practical workflows to guide researchers and drug development professionals in evaluating and integrating these complementary methodologies.
Quantitative Performance Benchmarking
A direct comparative analysis reveals the relative strengths and performance characteristics of traditional and AI/ML approaches under different conditions.
Table 1: Performance Comparison of NONMEM vs. AI/ML Models in PPK Analysis
| Model Category | Specific Model/Approach | Key Performance Metric (RMSE/MAE) | Computational Time | Key Strength | Primary Data Context | Source |
|---|---|---|---|---|---|---|
| Traditional NLMEM | NONMEM (FOCE, SAEM) | Baseline Reference | Variable (hrs-days) | Interpretability, Regulatory Acceptance | Real (n=1,770) & Simulated | [83] [32] |
| Machine Learning (ML) | XGBoost, Random Forest | Often outperformed NONMEM | Minutes-Hours | Handling non-linear covariate relationships | Real Clinical Datasets | [83] [84] |
| Deep Learning (DL) | Neural Networks (NN) | Often outperformed NONMEM | Moderate-High (GPU-dependent) | Pattern recognition in complex data | Large Real Clinical Datasets | [83] |
| Hybrid AI-NLMEM | Neural Ordinary Differential Equations (Neural ODE) | Strong performance, high explainability | High | Mechanistic + Data-driven fusion | Large Datasets | [83] |
| Automated PPK Search | pyDarwin (Bayesian Optimization) | Comparable to expert manual models | < 48 hours (avg., 40-CPU) | Exhaustive search, reproducibility | Synthetic & Real Clinical Datasets | [69] |
Interpretation of Benchmarks: The data indicates that AI/ML models, particularly neural ODEs and automated search algorithms, can match or exceed the predictive accuracy of traditional NONMEM models [83] [69]. A pivotal 2025 comparative study on real-world data from 1,770 patients found that AI/ML models "often outperform NONMEM," with neural ODEs providing a strong balance of performance and explainability [83]. Furthermore, automated AI-driven platforms can reliably identify optimal model structures in a fraction of the time required for manual development, significantly accelerating the popPK analysis timeline [69].
Detailed Experimental Protocols
Protocol 1: Traditional NONMEM Workflow for MM Parameter Estimation
This protocol outlines the established, iterative process for developing a population MM model using NONMEM.
Model Specification:
- Define the structural base model using the MM equation: ( dA/dt = -V{max} \cdot C / (Km + C) ), where ( V{max} ) is the maximum rate and ( Km }) is the substrate concentration at half-maximal velocity.
- Specify the statistical model for inter-individual variability (IIV) on parameters (e.g., exponential error model for ( V{max} ) and ( Km )), covariance (OMEGA matrix), and residual error model (SIGMA).
Parameter Estimation:
- Algorithm Selection: Choose an estimation method based on model complexity and data [32].
- FOCE with INTERACTION: Recommended for most PK/PD models, including MM, with moderate nonlinearity.
- Stochastic Approximation EM (SAEM): Preferred for complex models (e.g., with ODEs) or highly sparse data, as it provides a more accurate stochastic assessment of the parameter integral [32].
- Handling Censored Data (e.g., BLQ): For data below the limit of quantification, the Beal M3 method (likelihood-based) is precise but can be unstable. A pragmatic alternative is the M7+ method (impute zero, inflate additive error for BLQ points), which offers improved stability during model development with comparable bias and precision [63].
- Algorithm Selection: Choose an estimation method based on model complexity and data [32].
Model Diagnostics & Refinement:
- Evaluate goodness-of-fit plots: observations vs. population/individual predictions, conditional weighted residuals.
- Perform a visual predictive check (VPC) to assess simulation properties.
- Conduct stepwise covariate modeling (forward inclusion/backward elimination) using objective function value (OFV) changes.
Model Qualification:
- Finalize parameter estimates, compute confidence intervals via bootstrap or sampling importance resampling (SIR), and prepare model for simulation.
Protocol 2: AI/ML Model Development & Benchmarking Protocol
This protocol describes the process for developing and validating an AI/ML predictor, such as a neural network, for concentration or parameter prediction, tailored for comparison against a NONMEM MM model.
Problem Framing & Data Preparation:
- Define Task: Regression (predicting drug concentration) or classification (predicting dose group).
- Curate Dataset: Use the same dataset as the NONMEM analysis. Features may include time, dose, demographic covariates, genetic markers, and lab values [84].
- Preprocess: Handle missing values, scale/normalize features, and split data into training, validation, and test sets (e.g., 70/15/15). Ensure the test set is completely held out from training.
Model Selection & Training:
- Algorithm Choice: Based on [83] [84], select from:
- Gradient Boosting (XGBoost, LightGBM): Often high-performing with structured data.
- Neural Networks (Multilayer Perceptron): For capturing complex interactions.
- Neural ODE: To embed a mechanistic PK structure within a neural network framework [83].
- Training: Use the training set to fit the model. Employ K-fold cross-validation on the training set to tune hyperparameters (e.g., learning rate, network depth) and prevent overfitting [85].
- Algorithm Choice: Based on [83] [84], select from:
Benchmarking vs. NONMEM:
- Prediction: Generate predictions on the identical, unseen test set using the final trained AI/ML model and the final estimated NONMEM model.
- Performance Calculation: Compute and compare key metrics for both models:
- Root Mean Squared Error (RMSE)
- Mean Absolute Error (MAE)
- Coefficient of Determination (R²)
Explainability & Integration:
- Perform feature importance analysis (e.g., SHAP values for tree-based models) to interpret predictions.
- For hybrid approaches, consider using the AI model to inform covariate relationships or initial parameter estimates for a subsequent NONMEM run.
Visualizing the Hybrid AI-NONMEM Workflow
The integration of AI and traditional modeling is a powerful emerging paradigm. The following diagram illustrates a synergistic workflow where AI augments and informs the classic NONMEM modeling process.
Diagram 1: Integrated AI-NONMEM Workflow for Enhanced Model Development (Max Width: 760px)
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Software and Tools for Performance Benchmarking
| Tool Category | Specific Tool/Name | Primary Function in Research | Application Context |
|---|---|---|---|
| Traditional NLMEM Engine | NONMEM (ICON plc) | Gold-standard software for population PK/PD parameter estimation via NLMEM. | Core engine for developing traditional MM and other PK models [83] [32]. |
| AI/ML Modeling Frameworks | Python (Scikit-learn, PyTorch, TensorFlow) | Provides libraries for building, training, and validating a wide array of ML and DL models. | Implementing XGBoost, Neural Networks, Neural ODEs for predictive modeling [83] [84]. |
| Automated PopPK Search | pyDarwin | Implements global optimization algorithms (e.g., Bayesian Optimization) to automate popPK model structure search. | Rapid, reproducible identification of optimal model structures from a pre-defined space [69]. |
| LLM for Code Assistance | OpenAI o1 / GPT-4.1 | Generates accurate foundational NONMEM code for standard tasks, accelerating model coding. | Assisting in writing basic NONMEM control streams; requires expert review for complex models [86]. |
| Data Handling & Censoring | M7+ Method (Impute Zero, Inflate Error) | A pragmatic method for handling BLQ data, offering stability during model development. | Alternative to the more precise but less stable Beal M3 method in NONMEM [63]. |
The pursuit of precise Michaelis-Menten parameter estimation using NONMEM represents a cornerstone of modern pharmacometrics, providing a robust framework for understanding enzyme-mediated nonlinear pharmacokinetics. Within this established paradigm, a significant methodological evolution is underway with the emergence of Neural Ordinary Differential Equations (Neural ODEs). This hybrid approach synthesizes the principled, interpretable structure of differential equations with the adaptive, data-driven learning capacity of neural networks [87]. Traditional Nonlinear Mixed Effects (NLME) modeling, while powerful, operates under predefined structural assumptions and can be challenged by high-dimensional covariate relationships [88]. Neural ODEs offer a compelling alternative by learning the underlying system dynamics directly from observed data, using a neural network to parameterize the right-hand side of an ODE system [89] [90]. This fusion creates a tool that is not intended to replace mechanistic modeling but to augment it—particularly in scenarios where system dynamics are complex, partially unknown, or involve intricate covariate interactions that are difficult to specify a priori [91]. The integration of low-dimensional Neural ODE structures into pharmacometric software like Monolix and NONMEM marks a pivotal step toward practical application, enabling these models to account for inter-individual variability and serve as powerful support models within the traditional NLME workflow [92]. This document details the application notes and protocols for leveraging Neural ODEs within the context of advanced pharmacokinetic research.
Performance Benchmarking: Neural ODEs vs. Traditional PK Models
Empirical studies demonstrate the potential advantages of Neural ODEs, particularly in generalization and covariate integration. The following tables summarize key quantitative findings from recent applications.
Table 1: Cross-Regimen Generalization Performance with T-DM1 Data [93]
| Model | Training Regimen | Test Regimen | Key Performance Metric | Result |
|---|---|---|---|---|
| Neural ODE | Q3W (3-weekly) | Q1W (weekly) | Prediction Accuracy | Substantially better than ML/DL comparators |
| LSTM Network | Q3W (3-weekly) | Q1W (weekly) | Prediction Accuracy | Poor generalization to unseen regimen |
| LightGBM | Q3W (3-weekly) | Q1W (weekly) | Prediction Accuracy | Poor generalization to unseen regimen |
| Neural ODE | Q1W (weekly) | Q3W (3-weekly) | Prediction Accuracy | Substantially better than ML/DL comparators |
| Traditional PopPK | Q3W (3-weekly) | Q1W (weekly) | Extrapolation Capability | Reasonable, but model-specific |
Table 2: Predictive Accuracy on Sparse Dalbavancin PK Data (6-fold CV) [88] [94]
| Model Type | Covariate Handling | Mean Prediction Error | Relative Performance |
|---|---|---|---|
| Two-Compartment Model | Without Covariates | Baseline | Reference |
| NLME Model | With Covariate Search | Comparable to Baseline | Similar to compartment model |
| Neural ODE | Without Covariates | Comparable to Baseline | Matches state-of-the-art NLME |
| Neural ODE | With Integrated Covariates | Superior Accuracy | Best performance, leveraging complex covariate structures |
Table 3: NONMEM Estimation Methods for Complex Models [32]
| Estimation Method | Principle | Best For | Computational Cost |
|---|---|---|---|
| First Order (FO) | Linearizes inter & intra-subject variability | Simple PK models, initial estimates | Low |
| FOCE w/ Interaction | More accurate approximation of random effects | Standard for most PopPK/PD models | Moderate |
| Monte Carlo SAEM | Stochastic approximation of exact integral | Complex models (e.g., ODEs, >6 params) | High, but efficient for complex problems |
| Importance Sampling (IMP) | Monte Carlo integration around mode | Final evaluation, standard error calculation | High |
Core Experimental Protocol: Developing a Neural ODE PK Model
This protocol outlines the key steps for constructing and validating a Neural ODE model for pharmacokinetic data, integrating lessons from recent case studies [93] [92] [88].
3.1 Data Preparation and Preprocessing
- Data Structure: Organize data in a subject-centric format. Essential columns include: subject ID (
ID), time (TIME), drug concentration (DV), dosing amount (AMT), dosing interval/regimen indicator, and cycle number (CYCLE) [93]. For covariate modeling, append columns like age, weight, and biomarkers. - Feature Engineering for Time Series: For multi-cycle data, incorporate the first cycle's concentration profile (
PK_cycle1) as a predictive feature for subsequent cycles [93]. Handle values below the limit of quantification (BLQ) as per standard PK practices (e.g., omission or M3 method) [32]. - Data Augmentation for Sparse Data: To counteract overfitting with small datasets (N~200), employ data augmentation in pre-training. This can involve adding Gaussian noise to observed concentrations or simulating virtual subjects by perturbing covariates within plausible ranges [88].
3.2 Neural ODE Architecture Specification
- Low-Dimensional Network Design: Use a shallow network (e.g., one hidden layer with 5-20 neurons) to ensure identifiability and reduce overfitting, aligning with pharmacometric principles [87] [91]. A sample formulation for a one-dimensional state (concentration) is:
dh(t)/dt = f_NN(h(t), t, covariates; θ)wheref_NNis a neural network with ReLU activation andθare its weights [92] [91]. - Covariate Integration: Directly feed static covariates into the neural network alongside the state
h(t)and timet, allowing the model to learn complex, non-linear covariate-effect relationships directly from data [88]. - Mechanistic Hybridization: For partially known systems, combine a mechanistic ODE with a neural network component. For example, a known PK model structure can be used while a neural network approximates an unknown PD relationship or a complex absorption process [92] [87].
3.3 Model Training and Estimation
- Loss Function: Minimize the mean squared error (MSE) between observed and predicted concentrations [88] [90].
- Optimization in NONMEM/Monolix: When implemented as an NLME model, use the SAEM estimation algorithm for stability with complex ODEs [32] [92]. Increase the maximum number of iterations (e.g., to 1000). For parameters without inter-individual variability (IIV), test both the "no variability" and "variability at the first stage" estimation methods [92].
- Parameter Initialization: Crucial for convergence. Initialize neural network weights (e.g.,
W,b) such that neurons are not dead (e.g.,W*state + b > 0initially). Use software auto-initialization features followed by manual adjustment [92]. Initial estimates for traditional PK parameters (e.g.,CL,V) should follow standard practices.
3.4 Model Diagnostics and Validation
- Diagnostics: Evaluate standard goodness-of-fit plots: observed vs. population/individual predictions, conditional weighted residuals vs. time/predictions. Simulate from the fitted model to create Visual Predictive Checks (VPCs) [92].
- Interpretability Analysis: Employ SHapley Additive exPlanations (SHAP) to interpret how the Neural ODE uses input covariates for predictions, providing insight into learned covariate relationships [88] [94].
- Validation: Perform cross-validation (e.g., 6-fold) to assess robustness, especially with sparse data [88]. The ultimate test is external validation on a truly unseen dosing regimen to confirm superior generalizability [93].
Implementation Workflow: From NONMEM to a Hybrid Framework
Table 4: Software and Computational Toolkit
| Tool | Primary Use | Key Feature for Neural ODEs | Reference |
|---|---|---|---|
| NONMEM (v7.5+) | Industry-standard NLME modeling. | Supports ODEs; SAEM algorithm suitable for complex NODE estimation. | [32] [92] |
| Monolix Suite (2023R1+) | User-friendly NLME modeling. | Explicit tutorial for implementing low-dimensional Neural ODEs. | [92] |
| PyTorch / torchdiffeq | Flexible DL research in Python. | Provides differentiable ODE solvers for custom NODE development and prototyping. | [88] [90] |
| R (rxode2, torch) | Pharmacometrics & statistics. | Enables NODE prototyping and integration with statistical analysis pipelines. |
Table 5: Data and Modeling Components
| Component | Description | Protocol Consideration |
|---|---|---|
| Rich Time-Series PK Data | Multi-cycle, multi-regimen data (e.g., T-DM1 dataset). | Essential for testing model generalization across regimens [93]. |
| Patient Covariates | Demographics, lab values, biomarkers. | Feed directly into NODE to learn complex effect relationships [88]. |
| Low-Dim NN Architecture | Shallow network (1-2 hidden layers, <20 neurons). | Prevents overfitting on sparse PK data; aligns with PK principles [87] [91]. |
| SAEM Estimation Algorithm | Stochastic Approximation EM. | Preferred method in NONMEM/Monolix for stable NODE parameter estimation [32] [92]. |
| SHAP Analysis Library | SHapley Additive exPlanations. | Critical for interpreting the "black box" and understanding covariate influence [88]. |
This analysis evaluates key quantitative modeling paradigms—Nonlinear Mixed-Effects (NLME) modeling, Machine Learning (ML), and emerging hybrid and fractional approaches—within the specific context of Michaelis-Menten (M-M) enzyme kinetics and pharmacokinetic (PK) research. The central thesis frames NLME software like NONMEM as the historical and contemporary standard for reliable parameter estimation (e.g., Vmax, Km) in M-M systems [24]. However, the field is rapidly evolving with the introduction of data-hungry ML methods promising predictive power [95], mechanistic fractional models offering flexible dynamics [17], and Large Language Models (LLMs) entering as potential assistive tools [96]. The critical appraisal of these paradigms hinges on three pillars: interpretability (the mechanistic transparency of the model), extrapolation (performance beyond the conditions of the training data), and data requirements (the volume and quality of data needed for development and reliable application). Understanding these trade-offs is essential for selecting a fit-for-purpose model in drug development [43].
Table: Comparative Analysis of Modeling Paradigms in Pharmacometrics
| Paradigm | Core Strength | Key Limitation | Interpretability | Extrapolation Capability | Typical Data Requirements |
|---|---|---|---|---|---|
| NLME (NONMEM) | Robust, gold-standard for parameter estimation; quantifies population & individual variability [32]. | Can be computationally intensive; model building can be iterative and time-consuming. | High (Mechanistic parameters like CL, Vd, Vmax, Km). | Strong within the mechanistic domain of the model [43]. | Sparse to rich longitudinal data; handles unbalanced designs well. |
| Machine Learning (ML) | High predictive accuracy from complex, high-dimensional data; automates pattern discovery [95]. | "Black-box" nature; risk of false correlations; poor out-of-domain extrapolation. | Low (Correlative features, not causal mechanisms). | Generally poor (Data-driven, interpolative). | Very large datasets required for training and validation. |
| Hybrid (PPK-ML) | Augments PPK predictability with ML; can improve precision when data are limited [95]. | Complexity in integration; interpretability can be intermediate. | Medium to High (Depends on PPK core). | Improved over pure ML, guided by PPK structure. | Moderate to large; leverages PPK model to reduce pure ML data needs. |
| Fractional PK (FDEs) | Describes "anomalous" kinetics (e.g., power-law) with fewer parameters; memory effects [17]. | Novel mathematical framework; less familiar to researchers; specialized software needed. | Medium (Parameters lack classic physiological analogs). | Potentially strong within its mathematical domain. | Similar to classic NLME. |
| Large Language Models (LLMs) | Assistive tool for code generation, literature synthesis, and workflow automation [96]. | Hallucinations; lack of pharmacometrics-specific training data; not a standalone modeling tool. | Low for modeling, High for text tasks. | Not applicable for direct PK/PD prediction. | Not applicable (Used as tool, not a data-fitting model). |
NONMEM Estimation Methods for Michaelis-Menten Kinetics
Parameter estimation for the M-M equation (V = (Vmax * [S]) / (Km + [S])) is foundational. A simulation study comparing linearization methods (e.g., Lineweaver-Burk) to nonlinear estimation within NONMEM demonstrated the clear superiority of the nonlinear approach. When estimating Vmax and Km from in vitro elimination kinetic data, nonlinear methods provided the most accurate and precise results, particularly when data incorporated a realistic combined (additive + proportional) error model [24]. NONMEM offers a suite of estimation algorithms, each with specific utility for problems of varying complexity.
Table: Key NONMEM Estimation Methods for PK/PD Modeling
| Method (Guide) | Key Principle | Best Use Case | Relative Speed | Note on M-M Kinetics |
|---|---|---|---|---|
| First Order (FO) [32] | Linearizes inter-individual variability. | Initial screening, simple models. | Fast | May be inaccurate for highly nonlinear M-M. |
| FOCE [32] | Conditional estimation; more accurate approximation than FO. | Standard workhorse for most PK/PD models. | Moderate | Preferred over FO for M-M parameter estimation. |
| FOCE with INTERACTION [32] | Accounts for interaction between inter & intra-individual error. | Models with proportional or combined error structures. | Moderate | Crucial for M-M with combined error models [24]. |
| Laplace [32] | Second-order conditional estimation. | Non-normal data, highly nonlinear models. | Slower | Useful for complex PD or categorical data linked to PK. |
| IMP [32] | Monte Carlo Expectation Maximization (exact method). | Complex models (e.g., ODEs, >6 params), sparse data. | Slow (Stochastic) | Excellent for precise M-M estimation if computational cost is acceptable. |
| SAEM [32] | Stochastic Approximation EM (exact method). | Very complex models, population models with mixtures. | Slow (Stochastic) | Efficient for large problems; often used with IMP for final estimates. |
Detailed Experimental and Simulation Protocols
Protocol 1: Simulation Study for Comparing M-M Estimation Methods [24]
- Objective: To evaluate the accuracy and precision of Vmax and Km estimates from traditional linearization vs. nonlinear mixed-effects methods.
- Step 1 – Data Simulation:
- Use a pharmacokinetic simulation tool (e.g., R) to generate 1000 replicate datasets.
- Simulate substrate concentration (
[S]) over time based on a known M-M model with defined true Vmax and Km. - Incorporate realistic residual error: 1) Additive error model, and 2) Combined (additive + proportional) error model.
- Step 2 – Parameter Estimation:
- Apply multiple estimation methods to each replicate dataset.
- Linearization Methods: Include Lineweaver-Burk, Eadie-Hofstee, etc.
- Nonlinear Method: Implement using NONMEM (FOCE with INTERACTION recommended).
- Step 3 – Performance Evaluation:
- For each method and replicate, calculate the estimated Vmax and Km.
- Compute Relative Bias:
(Median of Estimates - True Value) / True Value. - Compute Precision: Use the 90% confidence interval of the estimates.
- Compare methods: The one with bias closest to zero and the narrowest confidence intervals is superior.
- Key Finding: Nonlinear estimation via NONMEM provided the most accurate and precise parameter recovery, especially under the combined error model [24].
Protocol 2: Developing a Hybrid PPK-ML Model for Vancomycin Dosing [95]
- Objective: To develop and compare a hybrid model against pure PPK and ML models for predicting vancomycin exposure (AUC24) in sepsis patients.
- Step 1 – Data Curation:
- Source electronic health record data (e.g., from MIMIC-IV).
- Include patient covariates (age, weight, serum creatinine), dosing records, and concentration measurements.
- Split data into a training set (for model development) and a test set (for validation).
- Step 2 – Base PPK Model Development:
- Use NONMEM to develop a population PK model (e.g., 1- or 2-compartment).
- Estimate typical population parameters (CL, V), inter-individual variability (IIV), and residual error.
- Perform covariate analysis (e.g., weight on CL, creatinine clearance on CL).
- Step 3 – Hybrid/ML Model Development:
- Extract empirical Bayes estimates (EBEs) of individual PK parameters (CLi, Vi) from the final NONMEM model.
- Use these EBEs as additional input features alongside original covariates in an ML algorithm (e.g., Random Forest, XGBoost).
- Train the ML model to predict drug concentration or directly AUC24.
- Step 4 – Validation & Comparison:
- Apply the standalone PPK model, the standalone ML model (trained without EBEs), and the hybrid model to the test set.
- Compare predictive performance using metrics: Mean Absolute Prediction Error (MAPE), Root Mean Squared Error (RMSE).
- Key Finding: The hybrid model often outperforms the pure PPK model when concentration data are unavailable, while Bayesian estimation (using the PPK model with concentration data) is best when concentrations are available [95].
Workflow for Model Evaluation and Stability
A critical challenge in pharmacometrics is ensuring model stability—where performance is reliable and not overly sensitive to small changes in data or settings. Instability manifests as convergence failures, unreasonable parameter estimates, or lack of standard errors [43]. The following diagnostic workflow, centered on NONMEM but applicable broadly, helps isolate the root cause, which typically lies in the balance between model complexity and data information content.
Workflow for Diagnosing Model Instability [43] A heuristic guide for diagnosing and resolving common model stability issues in pharmacometric analyses.
Workflow Narrative: The process begins by characterizing the nature of instability. If problems are systematic (e.g., consistently failing across runs), the issue likely relates to the fundamental mismatch between the chosen model's complexity and the information content provided by the experimental study design. For example, attempting to estimate a full target-mediated drug disposition (TMDD) model from data that only exhibits linear pharmacokinetics. The solution pathway involves model simplification, parameter fixing, or employing more robust estimation methods like IMP [43] [32].
Conversely, if instability is random or noisy (e.g., convergence depends on initial estimates), the root cause is more likely data quality or a model specification error. This pathway requires auditing data for outliers, verifying the residual error model, and meticulously checking the model code for non-syntactical errors (e.g., incorrect mapping of parameters) [43]. A stable model requires both a design that informs the parameters and high-quality data that aligns with the model's assumptions.
The Scientist's Toolkit: Essential Research Reagents and Software
Table: Key Research Reagent Solutions for PK/PD Modeling
| Item | Category | Function in Research | Example/Note |
|---|---|---|---|
| NONMEM | Primary Modeling Software | The industry standard for NLME model development, estimation, and simulation. | Used for M-M parameter estimation [24], PPK model development [95]. |
| R or Python | Statistical Computing & Scripting | Data preprocessing, visualization (ggplot2), post-processing of NONMEM outputs, running simulation studies. | Essential for implementing the simulation protocol in [24]. |
| Monolix | Alternative Modeling Software | NLME modeling software with a different estimation engine (SAEM) and user interface. | Used as an alternative to NONMEM in some research and industry settings. |
| Xpose / Pirana | Diagnostics Toolkit | R-based (Xpose) or standalone (Pirana) tool for diagnostic model evaluation and run management. | Facilitates goodness-of-fit plots and covariate model diagnostics. |
| PDx-Pop | Commercial Interface | A commercial interface for NONMEM, providing a workflow management and graphical environment. | Common in industry to streamline modeling workflows. |
| Simulation Dataset | Virtual Research Reagent | A rigorously constructed simulated dataset with known "true" parameters. | Critical for method comparison (as in [24]) and assessing model performance. |
| Benchmark PK Dataset | Empirical Research Reagent | A well-curated, public real-world dataset (e.g., from critical care databases). | Used for developing and testing models like the vancomycin hybrid model [95]. |
| Fractional Differential Equation Solver | Specialized Computational Tool | A numerical solver for implementing fractional PK models within an estimation framework. | Required for implementing the FDE approach described in [17]. |
The estimation of Michaelis-Menten parameters (V~max~ and K~m~) is a cornerstone of pharmacokinetics, critical for characterizing saturable metabolic and transport processes. While the underlying equation is well-established, the accuracy and precision of parameter estimation depend heavily on the chosen methodology and the software ecosystem that supports it [24]. Historically, researchers relied on linear transformation methods (e.g., Lineweaver-Burk, Eadie-Hofstee) which, while simple, can introduce significant bias and error propagation [24].
The advent of nonlinear mixed-effects modeling (NONMEM) software provided a paradigm shift, enabling direct fitting of the nonlinear Michaelis-Menten model to data. This approach, particularly within a population framework, allows for more robust estimation by accounting for inter-individual variability and complex residual error structures [24] [26]. The true power of NONMEM, however, is unlocked through its integration with a suite of auxiliary tools. This ecosystem, featuring PsN (Perl-speaks-NONMEM) for automation and advanced statistics, Pirana as a graphical workflow manager, and resources like NONMEMory for model libraries, has evolved into an indispensable platform for pharmacometric research [97] [98].
This integration directly addresses the findings of simulation studies which demonstrate that nonlinear estimation methods via NONMEM yield more reliable parameter estimates compared to traditional linearization techniques, especially with complex error models [24]. The evolution of this ecosystem—from standalone NONMEM execution to a connected, efficient workflow—forms the operational backbone of modern, model-informed drug development for Michaelis-Menten and other complex analyses.
Integrated Toolbox: Core Components and Their Synergistic Functions
The modern NONMEM-centric workflow is supported by specialized tools that manage, execute, and diagnose complex modeling tasks. Their roles are distinct but highly interconnected.
Pirana serves as the central graphical workbench and project manager. It provides a unified interface to organize model files, datasets, and output results. Its primary function is to streamline the execution of NONMEM runs through other tools in the ecosystem. For instance, it can launch PsN commands directly from its interface, manage remote computation hosts, and automatically refresh results upon job completion [99]. Recent versions have expanded its scope beyond NONMEM to integrate additional modeling engines like the NLME engine via R, and machine learning packages such as pyDarwin for automated model building [97].
PsN (Perl-speaks-NONMEM) is the computational engine for automation and advanced diagnostics. It is a command-line toolkit built on Perl modules that wraps around NONMEM to perform tasks impractical to code manually [98]. Its functions are critical for rigorous model evaluation and development:
- Automated Workflows: Tools like the stepwise covariate model (
scm) automate the tedious process of testing covariate relationships. - Model Diagnostics: Tools such as
vpc(Visual Predictive Check) andsimeval(simulation-based evaluation) are essential for assessing model predictive performance. - Robustness Assessment: Routines like
bootstrap(for confidence intervals) andcdd(case deletion diagnostics) evaluate the stability and reliability of parameter estimates [98].
NONMEM remains the core estimation engine. The latest version, NONMEM 7.6, includes advanced algorithms like Stochastic Approximation Expectation-Maximization (SAEM) and Markov-Chain Monte Carlo Bayesian Analysis (BAYES), which are particularly suited for complex nonlinear models [20]. Its NM-TRAN preprocessor allows models to be specified in a user-friendly control stream format.
The synergy is clear: A modeler uses Pirana to organize the project and then, with a click, launches a PsN vpc command. PsN automatically handles the simulation, NONMEM re-execution, and result compilation, with outputs neatly organized back into the Pirana interface for review [99] [97]. This seamless integration drastically reduces manual effort and potential for error.
Table 1: Core Software Components in the NONMEM Ecosystem for Michaelis-Menten Analysis
| Software | Primary Role | Key Function in Michaelis-Menten Context | Interface |
|---|---|---|---|
| NONMEM 7.6 | Gold-standard estimation engine [20] | Fits nonlinear Michaelis-Menten model using FOCE, SAEM, or other algorithms; estimates population V~max~ and K~m~ with variability. | Command-line, controlled via text files. |
| PsN (v5.6.0) | Automation & advanced statistics toolkit [98] | Automates VPC, bootstrap, covariate search, and model diagnostics to validate saturable PK model performance. | Command-line (executed directly or via Pirana). |
| Pirana | Modeling workbench & project manager [97] | Provides GUI to manage control streams, data, outputs; integrates and launches NONMEM/PsN; visualizes diagnostic plots. | Graphical User Interface (GUI). |
Application Notes & Protocols: Michaelis-Menten Parameter Estimation
Protocol: Simulation-Based Assessment of Estimation Methods
Objective: To compare the accuracy and precision of traditional linearization methods versus nonlinear mixed-effects estimation (using NONMEM) for determining V~max~ and K~m~ from in vitro drug elimination kinetic data [24].
Background: This protocol is based on a published simulation study by Cho et al. (2018), which demonstrated the superiority of nonlinear methods [24] [100]. The following is a detailed methodology for replicating and extending such an analysis using the integrated PsN/Pirana environment.
Materials & Software Requirements:
- Software: NONMEM (v7.4 or higher), PsN (v4.8 or higher), Pirana, R statistical software.
- Hardware: Computer with multi-core processor and ≥ 2 GB RAM dedicated to NONMEM [20].
Experimental/Simulation Procedure:
Data Simulation in R:
- Generate 1000 replicate datasets mimicking a typical in vitro incubation study.
- Use a prior V~max~ and K~m~ as population truths (e.g., from literature).
- Simulate substrate concentration ([S]) over time via numerical integration of the Michaelis-Menten ODE: -d[S]/dt = (V~max~ * [S]) / (K~m~ + [S]).
- Incorporate realistic residual error: either an additive error model (e.g., constant standard deviation) or a combined error model (constant + proportional), adding random noise via Monte Carlo simulation [24].
Model Development in Pirana/NONMEM:
- Control Stream Creation: Write a NONMEM control stream (
*.mod) defining the structural model.- Use
$PREDto code the integrated form of the Michaelis-Menten equation or use$DESwithADVAN13for differential equation solution. - Define parameters
VMAXandKMwith typical population values (THETA) and inter-individual variability (ETA) via an exponential model. - Define residual error (EPSILON) according to the simulation model (additive or combined).
- Use
- Dataset Formatting: Prepare the data file (
*.csv) with columns forID,TIME,DV(measured [S]),EVID, and potentiallyAMTif simulating dosing.
- Control Stream Creation: Write a NONMEM control stream (
Automated Execution with Pirana & PsN:
- Load the control stream and data file into a Pirana project.
- Use Pirana's "Execute" dialog (Ctrl-E) to run the base model. In the advanced view, select the PsN configuration and number of CPU nodes for parallel processing [99].
- The typical PsN command for a basic NONMEM run initiated from Pirana would be:
execute <model_name>.mod -dir=<run_directory>. - For the simulation study, this base estimation (using FOCE or SAEM) would be performed on each of the 1000 replicate datasets. This can be automated using a PsN script or a custom R loop that modifies the data file path for each run.
Statistical Analysis Plan:
- Parameter Estimation: For each replicate dataset, estimate parameters using:
- Traditional Methods: Linear transformations (Lineweaver-Burk, Eadie-Hofstee).
- NONMEM: Nonlinear mixed-effects estimation (FOCE method).
- Performance Metrics: For each method and each parameter (V~max~, K~m~), calculate:
- Relative Bias:
(Median Estimate - True Value) / True Value * 100% - Precision: Width of the 90% confidence interval (CI) for the estimates across replicates.
- Relative Bias:
- Comparison & Visualization: Use R (potentially via PsN's
rplotsoption [98]) to generate comparative boxplots of bias and tables of CI widths for all methods.
Expected Outcome: As demonstrated by Cho et al., the nonlinear estimation via NONMEM is expected to show significantly lower median bias and tighter confidence intervals, particularly for data generated with a combined error model, confirming its superior reliability [24].
Table 2: Performance Comparison of Estimation Methods for Michaelis-Menten Parameters (Simulated Data)
| Estimation Method | V~max~ Relative Bias (%) | K~m~ Relative Bias (%) | Remarks / Key Advantage |
|---|---|---|---|
| Lineweaver-Burk (Linear) | High Positive Bias [24] | High Negative Bias [24] | Simple but statistically flawed; distorts error structure. |
| Eadie-Hofstee (Linear) | Moderate Bias [24] | Moderate Bias [24] | Better than Lineweaver-Burk but still suboptimal. |
| NONMEM (FOCE) | Lowest Bias [24] | Lowest Bias [24] | Directly models nonlinearity and correct error structure; handles population data. |
| NONMEM (SAEM) | Theoretically Lower Bias | Theoretically Lower Bias | More robust for complex models or sparse data; available in NONMEM 7.6 [20]. |
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Essential Software Tools & Resources for Michaelis-Menten PK/PD Research
| Tool/Resource Name | Category | Primary Function | Application in Michaelis-Menten Context |
|---|---|---|---|
| NONMEM 7.6 [20] | Estimation Engine | Nonlinear mixed-effects modeling and simulation. | Gold-standard for population estimation of V~max~ and K~m~ with inter-individual variability. |
| PsN Toolkit [98] | Automation & Diagnostics | Perl-based modules for automating NONMEM runs and advanced statistics. | Automates VPC, bootstrap, and covariate search for saturable PK models. |
| Pirana [97] | Workbench / GUI | Integrated modeling environment for managing runs, outputs, and diagnostics. | Central hub for project management, visual diagnostics, and executing NONMEM/PsN workflows. |
| R / nlmixr2 Ecosystem [101] | Complementary Framework | Open-source platform for pharmacometrics; includes nlmixr2 for NLME modeling. |
Alternative estimation environment; packages like ggPMX and vpc provide diagnostic plots compatible with NONMEM outputs. |
| Xpose / ggPMX | Diagnostic Visualization | R packages for standardized goodness-of-fit and diagnostic plotting. | Generates publication-quality plots (e.g., predictions vs. observations, residual diagnostics) for model evaluation [101]. |
| Model Libraries (NONMEMory / nlmixr2lib) | Knowledge Repository | Community-shared collections of canonical models. | Provides templates and starting points for coding Michaelis-Menten elimination or absorption models [101]. |
| Pharmpy | Model Conversion & Toolbox | Python toolkit for pharmacometric analysis. | Can assist in converting models between formats (e.g., NONMEM to nlmixr2) and in automated model building [101]. |
Advanced Applications and Future Directions
The integrated ecosystem is expanding beyond traditional pharmacokinetic modeling. Pirana now integrates machine learning packages like pyDarwin, which can automatically search vast spaces of model structures, random effects, and covariate relationships [97]. This is particularly valuable for complex saturable systems where the standard Michaelis-Menten model may need extension (e.g., incorporating auto-inhibition or time-dependent degradation).
Furthermore, the rise of the open-source nlmixr2 ecosystem in R represents a parallel and interoperable evolution. It includes tools for automatic model initialization (nlmixr2autoinit), simulation (Campsis), and optimal design (PopED) [101]. Crucially, conversion tools like babelmixr2 and pharmpy allow models to be translated between NONMEM and nlmixr2 formats, fostering collaboration and method comparison [101].
Future directions, as highlighted by surveys in quantitative systems pharmacology (QSP), point toward a need for even better scripting capabilities, high-performance computing integration, and enhanced visualization [102]. These needs are being met through improved parallel computing in NONMEM 7.6 [20], cloud-based execution options, and the continuous development of R and Python packages for advanced diagnostics and reporting. The enduring goal is a cohesive, efficient, and transparent software environment that accelerates the translation of complex kinetic models, like Michaelis-Menten systems, into actionable drug development decisions.
Conclusion
The reliable estimation of Michaelis-Menten parameters using NONMEM remains a cornerstone for characterizing nonlinear pharmacokinetics in drug development. This guide has synthesized the journey from foundational theory and practical implementation to resolving instability and validating outcomes. The future of this field lies in the strategic integration of traditional, robust NLME methods with emerging technologies. This includes adopting automated pipelines for generating initial estimates to improve efficiency and reproducibility [citation:3], leveraging AI/ML models for specific predictive tasks where they excel [citation:2], and utilizing advanced algorithms within NONMEM itself for complex optimal dosing challenges [citation:4]. As these tools converge, pharmacometricians will be empowered to build more stable, predictive models faster, ultimately accelerating the delivery of safe and effective therapies, especially for drugs exhibiting saturable, target-mediated disposition.
References
- 1. Michaelis–Menten kinetics [https://en.wikipedia.org/wiki/Michaelis%E2%80%93Menten_kinetics]
- 2. Michaelis Menten Kinetics - an overview [https://www.sciencedirect.com/topics/immunology-and-microbiology/michaelis-menten-kinetics]
- 3. Michaelis-Menten Kinetics [https://chem.libretexts.org/Bookshelves/Biological_Chemistry/Supplemental_Modules_(Biological_Chemistry)/Enzymes/Enzymatic_Kinetics/Michaelis-Menten_Kinetics]
- 4. Comparison of various estimation methods ... [https://synapse.koreamed.org/articles/1094566]
- 5. Application of modified Michaelis – Menten equations for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8507113/]
- 6. Michaelis-Menten equation – Knowledge and References [https://taylorandfrancis.com/knowledge/Medicine_and_healthcare/Medical_genetics/Michaelis-Menten_equation/]
- 7. An automated pipeline to generate initial estimates for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12592298/]
- 8. R-based reproduction of the estimation process hidden ... [https://www.semanticscholar.org/paper/R-based-reproduction-of-the-estimation-process-%C2%AE-2-Bae-Yim/c3f78d95d7c999f2ecc789992a68a57c096f3c2c]
- 9. Drug Elimination - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/drug-elimination]
- 10. Comparison of various estimation methods for the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6989224/]
- 11. Advancing drug development with “Fit-for-Purpose” modeling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12436579/]
- 12. Pharmacokinetics - StatPearls - NCBI Bookshelf - NIH [https://www.ncbi.nlm.nih.gov/books/NBK557744/]
- 13. Comparison of various estimation [https://tcpharm.org/DOIx.php?id=10.12793/tcp.2018.26.1.39]
- 14. Machine Learning for PK/PD Modeling in Drug Development [https://www.certara.com/blog/machine-learning-for-pk-pd-modeling-in-drug-development/]
- 15. Saturable Drug Absorption [https://www.certara.com/knowledge-base/saturable-drug-absorption/]
- 16. Statistical identifiability and convergence evaluation for ... [https://www.sciencedirect.com/science/article/abs/pii/S0169260713003313]
- 17. Implementation of non-linear mixed effects models defined ... [https://link.springer.com/article/10.1007/s10928-023-09851-1]
- 18. Chapter 1 - Introduction to NONMEM, PREDPP, and NM-TRAN [https://nmhelp.tingjieguo.com/V/1]
- 19. NONMEM Tutorial Part I: Description of Commands and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6709426/]
- 20. NONMEM | Nonlinear Mixed Effects Modelling [https://www.iconplc.com/solutions/technologies/nonmem]
- 21. NONMEM [https://en.wikipedia.org/wiki/NONMEM]
- 22. NONMEM Code [https://www.pharmacy.umaryland.edu/centers/ctm/learning-resources/nonmem-code/]
- 23. Basic NONMEM Workshop Instructions [https://holford.fmhs.auckland.ac.nz/teaching/PAWS/workshops/basic/instructions]
- 24. Comparison of various estimation methods for the ... [https://pubmed.ncbi.nlm.nih.gov/32055546/]
- 25. Tutorial for Modeling Delays in Biological Systems in the ... [https://pubmed.ncbi.nlm.nih.gov/40524365/]
- 26. Evaluation of the nonparametric estimation method in ... [https://www.sciencedirect.com/science/article/abs/pii/S0928098708005125]
- 27. Introduction to Pharmacokinetic Modeling Methods - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3636497/]
- 28. Basic Concepts in Population Modeling, Simulation, and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3606044/]
- 29. Two NONMEM Tutorials in PSP! [https://www.ascpt.org/Journals/PSP/PSPost/View/ArticleId/24063/Two-NONMEM-Tutorials-in-PSP]
- 30. Chapter 4 - Models for Population Data - NONMEM Help [https://nmhelp.tingjieguo.com/V/4]
- 31. A Bayesian Approach for Population Pharmacokinetic ... [https://www.sciencedirect.com/science/article/pii/S0011393X1630131X]
- 32. NONMEM Tutorial Part II: Estimation Methods and Advanced ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6709422/]
- 33. Beginners Tutorial [https://www.pharmacy.umaryland.edu/centers/ctm/learning-resources/beginners-tutorial/]
- 34. Support software and resources [https://www.page-meeting.org/about-us/support-software-and-resources/]
- 35. NONMEM and R code for PKPD [https://pkpd-info.com/NONMEM.php]
- 36. I. Overview of PREDPP - NONMEM Help [https://nmhelp.tingjieguo.com/VI/I]
- 37. advan.ppp [https://pkpd-info.com/NONMEM/NM_guides/advan.htm]
- 38. User-Written Models [https://basicmedicalkey.com/user-written-models/]
- 39. des.for [https://pkpd-info.com/NONMEM/NM_guides/des.htm]
- 40. Chapter 7 - $SUBROUTINE Record and $PK Record [https://nmhelp.tingjieguo.com/V/7]
- 41. NONMEM Users Guide Part VI - PREDPP - Chapter I [https://nonmem.iconplc.com/nonmem/Nonmem_7_Project/nmvi2.0beta/htmlplus/VI/I.html]
- 42. An automated pipeline to generate initial estimates for ... [https://pubmed.ncbi.nlm.nih.gov/41199105/]
- 43. A Pharmacometric Workflow for Resolving Model Instability in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12521063/]
- 44. The impact of misspecified covariate models on inclusion ... [https://link.springer.com/article/10.1007/s10928-025-09964-9]
- 45. Chapter 6 - NONMEM Help [https://nmhelp.tingjieguo.com/V/6]
- 46. Interpreting the NONMEM Output [https://basicmedicalkey.com/interpreting-the-nonmem-output/]
- 47. A modeller's perspective on EM methods versus FOCE ... [https://www.page-meeting.org/default.asp?abstract=11725]
- 48. Comparing the performance of first-order conditional ... [https://pubmed.ncbi.nlm.nih.gov/33884580/]
- 49. Estimation and Solvers - NONMEM - Viktor Rognås [https://vrognas.com/docs/tools-of-the-trade/nonmem/nm-est/]
- 50. Performance comparison of first-order conditional estimation ... [https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/s12874-017-0427-0]
- 51. NONMEM PK Templates [https://pmxrepo.com/nmcode.html]
- 52. Bayesian estimation in NONMEM - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC10864934/]
- 53. Tutorial for Modeling Delays in Biological Systems in the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12256671/]
- 54. Tutorial for $DESIGN in NONMEM: Clinical trial evaluation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8674001/]
- 55. Leveraging large language models in pharmacometrics [https://pubmed.ncbi.nlm.nih.gov/40464844/]
- 56. NONMEM model templates and Codes [https://pkpd-info.com/NONMEM/Model_templates.php]
- 57. Modelling pulsatile profiles in NONMEM [https://www.pmxsolutions.com/2019/10/09/modelling-pulsatile-profiles-in-nonmem-2/]
- 58. OptiDose: Computing the Individualized Optimal Drug Dosing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8550736/]
- 59. Strategies for Dose Selection in Oncology Drug Development [https://www.caidya.com/strategies-for-dose-selection/]
- 60. Project Optimus [https://www.fda.gov/about-fda/oncology-center-excellence/project-optimus]
- 61. Computing optimal drug dosing regarding efficacy and safety [https://link.springer.com/article/10.1007/s10928-024-09940-9]
- 62. NONMEM on Metworx [https://kb.metworx.com/Users/Tutorials/Nonmem/Nonmem-on-Metworx/]
- 63. A Pragmatic Approach to Handling Censored Data Below ... [https://pubmed.ncbi.nlm.nih.gov/40067154/]
- 64. An automated pipeline to generate initial estimates for ... [https://link.springer.com/article/10.1007/s10928-025-10000-z]
- 65. NONMEM Overview and Writing an NM-TRAN Control ... [https://basicmedicalkey.com/nonmem-overview-and-writing-an-nm-tran-control-stream/]
- 66. 1. What This Chapter is About [https://nonmem.iconplc.com/nonmem/Nonmem_7_Project/nmvi2.0beta/htmlplus/V/11.html]
- 67. Introduction to Version VI - NONMEM Help [https://nmhelp.tingjieguo.com/intro/I]
- 68. A genetic algorithm based global search strategy for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4294074/]
- 69. A machine learning approach to population ... [https://www.nature.com/articles/s43856-025-01054-8]
- 70. An Evolutionary Search Algorithm for Covariate Models in ... [https://www.sciencedirect.com/science/article/abs/pii/S0022354917302642]
- 71. An automated pipeline to generate initial estimates for population ... [http://47.251.13.51:8081/41199105/]
- 72. Automated Covariate Model Building Within NONMEM [https://link.springer.com/article/10.1023/A:1011970125687]
- 73. Help for package nlmixr2autoinit [https://cran.r-project.org/web/packages/nlmixr2autoinit/refman/nlmixr2autoinit.html]
- 74. NONMEMory: A run management tool for NONMEM [https://www.sciencedirect.com/science/article/abs/pii/S0169260705000581]
- 75. Visual predictive check | Monolix - MonolixSuite Documentation [https://monolixsuite.slp-software.com/monolix/2024R1/visual-predictive-check]
- 76. A Tutorial on Visual Predictive Checks [https://www.page-meeting.org/Abstracts/a-tutorial-on-visual-predictive-checks/]
- 77. A step-by-step guide to prediction corrected visual ... [https://www.pmxsolutions.com/2018/09/21/a-step-by-step-guide-to-prediction-corrected-visual-predictive-checks-vpc-of-nonmem-models/]
- 78. Visual Predictive Checks (VPC) • xpose - GitHub Pages [https://uupharmacometrics.github.io/xpose/articles/vpc.html]
- 79. A Regression Approach to Visual Predictive Checks for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6202468/]
- 80. The Reference-Corrected Visual Predictive Check: A More ... [https://pubmed.ncbi.nlm.nih.gov/40299161/]
- 81. NONMEM's SIGDIG Convergence Criterion [https://holford.fmhs.auckland.ac.nz/research/sigdig]
- 82. Visual Predictive Checks (VPC) [https://cran.r-project.org/web/packages/xpose/vignettes/vpc.html]
- 83. A Comparative Analysis of NONMEM and AI-based Models ... [https://pubmed.ncbi.nlm.nih.gov/41254220/]
- 84. Artificial Intelligence and Machine Learning Applications to ... [https://www.mdpi.com/2079-6382/13/12/1203]
- 85. AI‐NLME: A New Artificial Intelligence‐Driven Nonlinear ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12405737/]
- 86. AI for NONMEM Coding in Pharmacometrics Research and ... [https://pubmed.ncbi.nlm.nih.gov/41187068/]
- 87. Low-dimensional neural ODEs and their application in ... [https://link.springer.com/article/10.1007/s10928-023-09886-4]
- 88. Leveraging Neural ODEs for Population Pharmacokinetics of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12192077/]
- 89. Understanding Neural ODE's - Jonty Sinai [https://jontysinai.github.io/jekyll/update/2019/01/18/understanding-neural-odes.html]
- 90. Chapter 3: Neural Ordinary Differential Equations [http://implicit-layers-tutorial.org/neural_odes/]
- 91. Low-dimensional neural ODEs and their application in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10982100/]
- 92. Using neural ODEs in Monolix | Tutorials [https://monolixsuite.slp-software.com/tutorials/2024R1/case-study-neural-odes]
- 93. Neural-ODE for pharmacokinetics modeling and its advantage ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8283337/]
- 94. Leveraging Neural ODEs for Population Pharmacokinetics ... [https://www.mdpi.com/1099-4300/27/6/602]
- 95. Machine learning and population pharmacokinetics [https://pmc.ncbi.nlm.nih.gov/articles/PMC12054080/]
- 96. Pharmacometrics in the Age of Large Language Models [https://pmc.ncbi.nlm.nih.gov/articles/PMC12567310/]
- 97. Pirana and Integrated PMX Tools, a Workbench for ... [https://pubmed.ncbi.nlm.nih.gov/40613737/]
- 98. PsN - Perl-speaks-NONMEM [https://uupharmacometrics.github.io/PsN/]
- 99. Using_NONMEM_through_PsN [https://onlinehelp.certara.com/pirana/23.10.1/Pirana_User_Guide/Pirana_NONMEM/Using_NONMEM_through_PsN.htm]
- 100. [PDF] Comparison of various estimation methods for the ... [https://www.semanticscholar.org/paper/Comparison-of-various-estimation-methods-for-the-of-Cho-Lim/0c92de62e2a1376fdd2cb4a642cd1b126953f081]
- 101. nlmixr2 ecosystem [https://blog.nlmixr2.org/blog/2025-06-30-nlmixr2-ecosystem/]
- 102. A Survey of Software Tool Utilization and Capabilities for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6389347/]